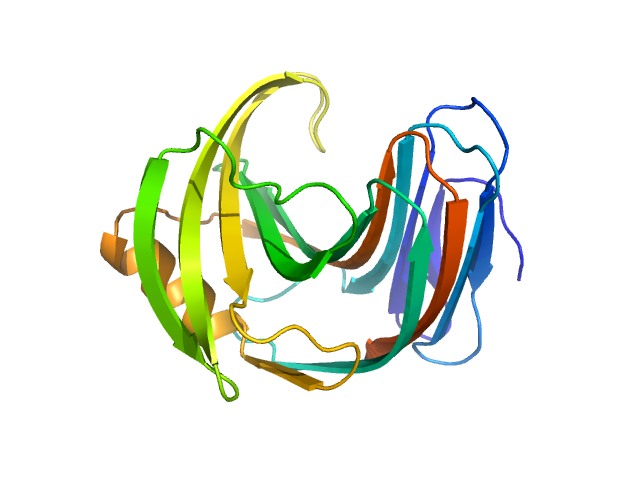
UniProt ID: F8W669 (1-190) Endo-1,4-beta-xylanase
|
|
|
|
| Sample: |
Endo-1,4-beta-xylanase monomer, 21 kDa Trichoderma longibrachiatum protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.2 |
nm |
|
|
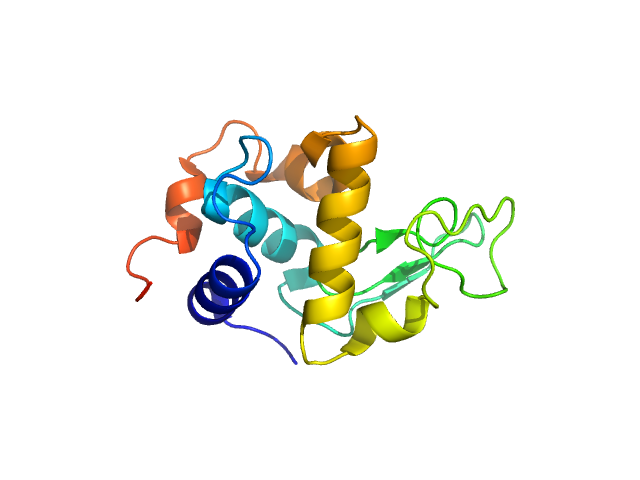
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
50 mM sodium citrate, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.7 |
nm |
|
|
UniProt ID: P42224 (1-750) Signal transducer and activator of transcription 1-alpha/beta
|
|
|
|
| Sample: |
Signal transducer and activator of transcription 1-alpha/beta tetramer, 349 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES-NaOH, 150 mM NaCl, 3% glycerol, 2 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Dec 11
|
Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein
Science Signaling 18(878) (2025)
Sugiyama A, Minami M, Ugajin K, Inaba-Inoue S, Yabuno N, Takekawa Y, Xiaomei S, Takei S, Sasaki M, Nomai T, Jiang X, Kita S, Maenaka K, Hirose M, Yao M, Gooley P, Moseley G, Sugita Y, Ose T
|
| RgGuinier |
6.2 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
800 |
nm3 |
|
|
UniProt ID: P42224 (1-750) Signal transducer and activator of transcription 1-alpha/beta
UniProt ID: Q9IPJ8 (1-297) Phosphoprotein
|
|
|
|
| Sample: |
Signal transducer and activator of transcription 1-alpha/beta tetramer, 349 kDa Homo sapiens protein
Phosphoprotein monomer, 33 kDa Rabies virus (strain … protein
|
| Buffer: |
10 mM HEPES-NaOH, 150 mM NaCl, 3% glycerol, 2 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Dec 11
|
Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein
Science Signaling 18(878) (2025)
Sugiyama A, Minami M, Ugajin K, Inaba-Inoue S, Yabuno N, Takekawa Y, Xiaomei S, Takei S, Sasaki M, Nomai T, Jiang X, Kita S, Maenaka K, Hirose M, Yao M, Gooley P, Moseley G, Sugita Y, Ose T
|
| RgGuinier |
6.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
857 |
nm3 |
|
|
UniProt ID: P42224 (1-750) Signal transducer and activator of transcription 1-alpha/beta
UniProt ID: None (None-None) Signal transducer and activator of transcription 1 binding DNA
|
|
|
|
| Sample: |
Signal transducer and activator of transcription 1-alpha/beta tetramer, 349 kDa Homo sapiens protein
Signal transducer and activator of transcription 1 binding DNA dimer, 11 kDa Homo sapiens DNA
|
| Buffer: |
10 mM HEPES-NaOH, 150 mM NaCl, 3% glycerol, 2 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Dec 11
|
Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein
Science Signaling 18(878) (2025)
Sugiyama A, Minami M, Ugajin K, Inaba-Inoue S, Yabuno N, Takekawa Y, Xiaomei S, Takei S, Sasaki M, Nomai T, Jiang X, Kita S, Maenaka K, Hirose M, Yao M, Gooley P, Moseley G, Sugita Y, Ose T
|
| RgGuinier |
6.1 |
nm |
| Dmax |
19.4 |
nm |
| VolumePorod |
761 |
nm3 |
|
|
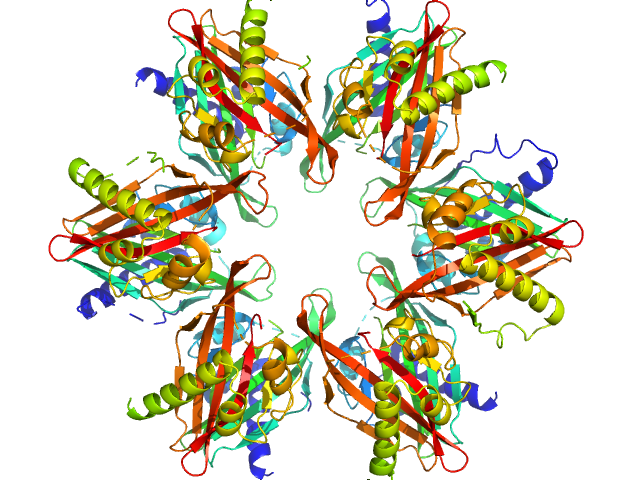
UniProt ID: Q9UQM7 (345-475) Calcium/calmodulin-dependent protein kinase type II subunit alpha
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha dodecamer, 186 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 150 mM NaCl, 1 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 3
|
Ligand‐induced CaMKIIα
hub Trp403 flip, hub domain stacking, and modulation of kinase activity
Protein Science 33(10) (2024)
Narayanan D, Larsen A, Gauger S, Adafia R, Hammershøi R, Hamborg L, Bruus‐Jensen J, Griem‐Krey N, Gee C, Frølund B, Stratton M, Kuriyan J, Kastrup J, Langkilde A, Wellendorph P, Solbak S
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
367 |
nm3 |
|
|
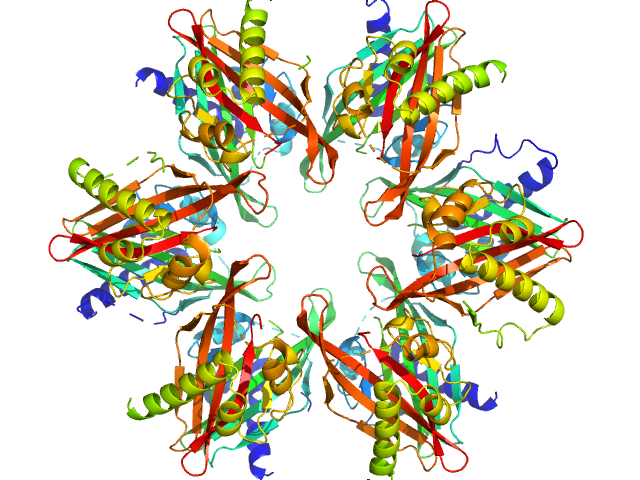
UniProt ID: Q9UQM7 (345-475) Calcium/calmodulin-dependent protein kinase type II subunit alpha
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha dodecamer, 186 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 150 mM NaCl, 1 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2020 Nov 19
|
Ligand‐induced CaMKIIα
hub Trp403 flip, hub domain stacking, and modulation of kinase activity
Protein Science 33(10) (2024)
Narayanan D, Larsen A, Gauger S, Adafia R, Hammershøi R, Hamborg L, Bruus‐Jensen J, Griem‐Krey N, Gee C, Frølund B, Stratton M, Kuriyan J, Kastrup J, Langkilde A, Wellendorph P, Solbak S
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
351 |
nm3 |
|
|
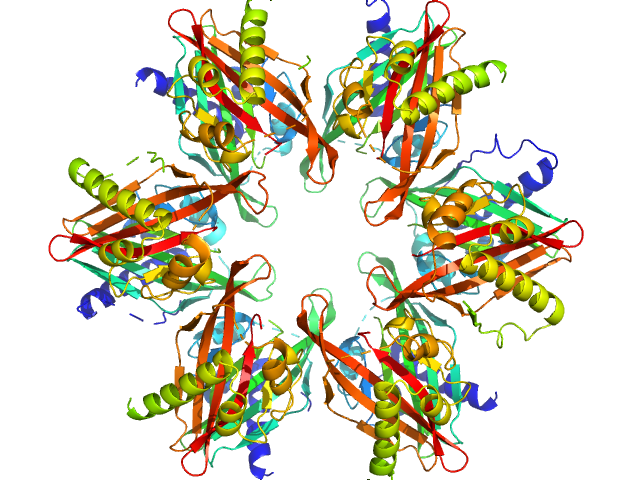
UniProt ID: Q9UQM7 (345-475) Calcium/calmodulin-dependent protein kinase type II subunit alpha
UniProt ID: None (None-None) 2-(6-(4-chlorophenyl)imidazo[1,2-b]pyridazine-2-yl)acetic acid
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha dodecamer, 186 kDa Homo sapiens protein
2-(6-(4-chlorophenyl)imidazo[1,2-b]pyridazine-2-yl)acetic acid monomer, 0 kDa
|
| Buffer: |
20 mM MES, 150 mM NaCl, 1 mM DTT, 200 µM PIPA, pH: 6 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2022 Nov 30
|
Ligand‐induced CaMKIIα
hub Trp403 flip, hub domain stacking, and modulation of kinase activity
Protein Science 33(10) (2024)
Narayanan D, Larsen A, Gauger S, Adafia R, Hammershøi R, Hamborg L, Bruus‐Jensen J, Griem‐Krey N, Gee C, Frølund B, Stratton M, Kuriyan J, Kastrup J, Langkilde A, Wellendorph P, Solbak S
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
396 |
nm3 |
|
|
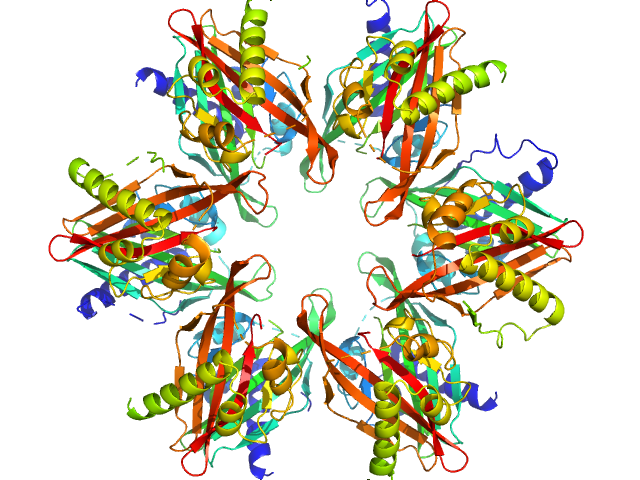
UniProt ID: Q9UQM7 (345-475) Calcium/calmodulin-dependent protein kinase type II subunit alpha
UniProt ID: None (None-None) 2-(6-(4-chlorophenyl)imidazo[1,2-b]pyridazine-2-yl)acetic acid
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha dodecamer, 186 kDa Homo sapiens protein
2-(6-(4-chlorophenyl)imidazo[1,2-b]pyridazine-2-yl)acetic acid monomer, 0 kDa
|
| Buffer: |
20 mM MES, 150 mM NaCl, 1 mM DTT, 200 µM PIPA, pH: 6 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2023 May 30
|
Ligand‐induced CaMKIIα
hub Trp403 flip, hub domain stacking, and modulation of kinase activity
Protein Science 33(10) (2024)
Narayanan D, Larsen A, Gauger S, Adafia R, Hammershøi R, Hamborg L, Bruus‐Jensen J, Griem‐Krey N, Gee C, Frølund B, Stratton M, Kuriyan J, Kastrup J, Langkilde A, Wellendorph P, Solbak S
|
| RgGuinier |
5.5 |
nm |
| Dmax |
27.0 |
nm |
|
|
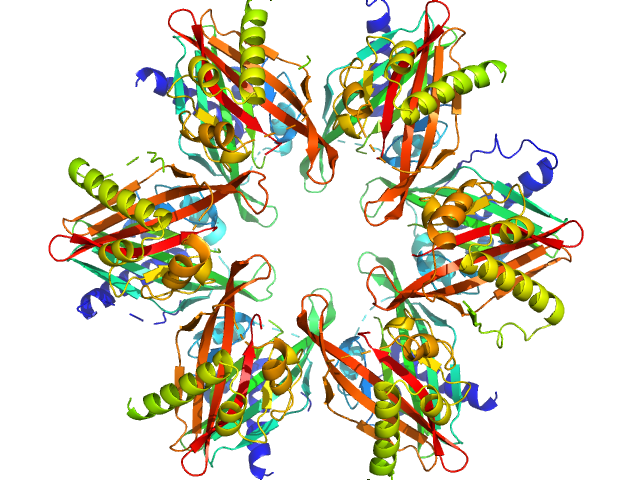
UniProt ID: Q9UQM7 (345-475) Calcium/calmodulin-dependent protein kinase type II subunit alpha
UniProt ID: None (None-None) 2-(6-(4-chlorophenyl)imidazo[1,2-b]pyridazine-2-yl)acetic acid
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha dodecamer, 186 kDa Homo sapiens protein
2-(6-(4-chlorophenyl)imidazo[1,2-b]pyridazine-2-yl)acetic acid monomer, 0 kDa
|
| Buffer: |
20 mM MES, 150 mM NaCl, 1 mM DTT, 200 µM PIPA, pH: 6 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2023 May 31
|
Ligand‐induced CaMKIIα
hub Trp403 flip, hub domain stacking, and modulation of kinase activity
Protein Science 33(10) (2024)
Narayanan D, Larsen A, Gauger S, Adafia R, Hammershøi R, Hamborg L, Bruus‐Jensen J, Griem‐Krey N, Gee C, Frølund B, Stratton M, Kuriyan J, Kastrup J, Langkilde A, Wellendorph P, Solbak S
|
| RgGuinier |
8.8 |
nm |
| Dmax |
37.0 |
nm |
|
|