UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 18-mer, 751 kDa Gallus gallus protein
|
| Buffer: |
50 mM KCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM ATP, 0.1% 2-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 18-mer, 751 kDa Gallus gallus protein
|
| Buffer: |
50 mM KCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM ATP, 0.1% 2-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 18-mer, 751 kDa Gallus gallus protein
|
| Buffer: |
50 mM KCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM ATP, 0.1% 2-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
4.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
5.2 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
266 |
nm3 |
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|

|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
4.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
241 |
nm3 |
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|

|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 1
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
4.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
243 |
nm3 |
|
|
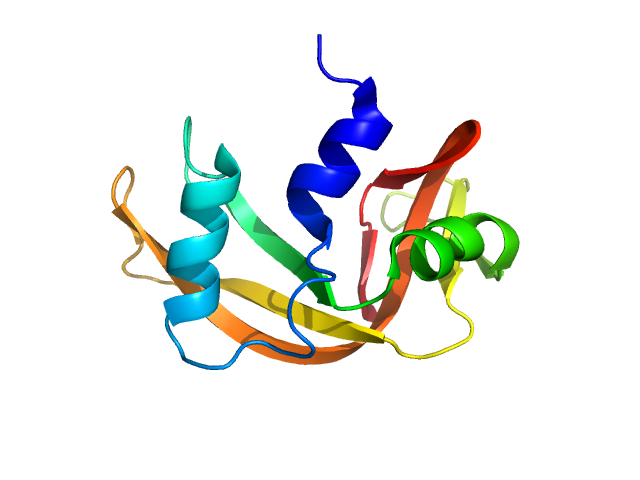
UniProt ID: P61823 (27-150) Ribonuclease pancreatic
|
|
|
|
| Sample: |
Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
|
|
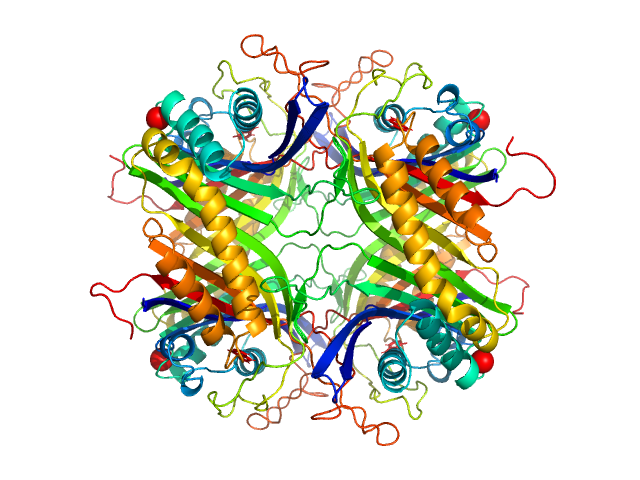
UniProt ID: Q00511 (2-302) Uricase
|
|
|
|
| Sample: |
Uricase tetramer, 136 kDa Aspergillus flavus protein
|
| Buffer: |
100 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.3 |
nm |
|
|
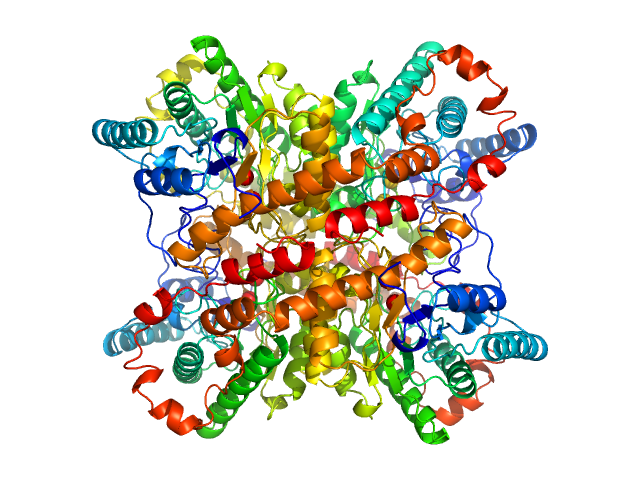
UniProt ID: P24300 (1-388) Xylose isomerase
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.1 |
nm |
|
|