UniProt ID: Q9UQM7 (345-475) Calcium/calmodulin-dependent protein kinase type II subunit alpha (T354N, E355Q, T412N, I414M, I464H, F467M)
UniProt ID: None (None-None) 5-hydroxydiclofenac
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha (T354N, E355Q, T412N, I414M, I464H, F467M) 14-mer, 217 kDa Homo sapiens protein
5-hydroxydiclofenac monomer, 0 kDa
|
| Buffer: |
20 mM MES, 150 mM NaCl, 1 mM DTT, 100 µM 5-HDC, pH: 6 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2020 Nov 19
|
Ligand‐induced CaMKIIα
hub Trp403 flip, hub domain stacking, and modulation of kinase activity
Protein Science 33(10) (2024)
Narayanan D, Larsen A, Gauger S, Adafia R, Hammershøi R, Hamborg L, Bruus‐Jensen J, Griem‐Krey N, Gee C, Frølund B, Stratton M, Kuriyan J, Kastrup J, Langkilde A, Wellendorph P, Solbak S
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
366 |
nm3 |
|
|
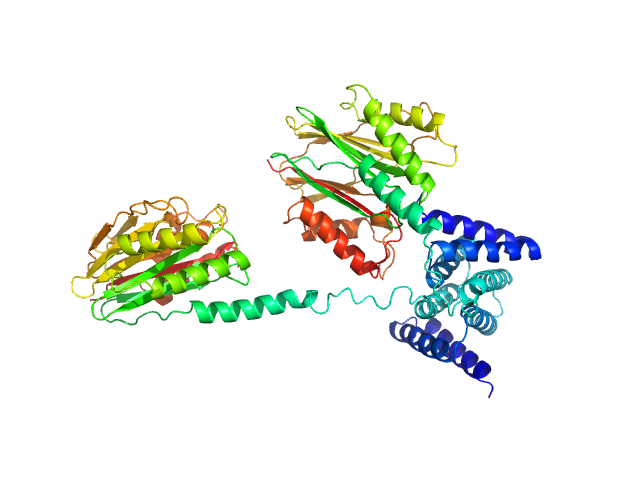
UniProt ID: P40399 (1-335) Phosphoserine phosphatase RsbU
|
|
|
|
| Sample: |
Phosphoserine phosphatase RsbU dimer, 77 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 24
|
A general mechanism for initiating the bacterial general stress response.
Elife 13 (2025)
Baral R, Ho K, Kumar RP, Hopkins JB, Watkins MB, LaRussa S, Caban-Penix S, Calderone LA, Bradshaw N
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
UniProt ID: P42411 (1-133) Serine/threonine-protein kinase RsbT
UniProt ID: P40399 (1-335) Phosphoserine phosphatase RsbU (Q94L)
|
|
|
|
| Sample: |
Serine/threonine-protein kinase RsbT dimer, 29 kDa Bacillus subtilis (strain … protein
Phosphoserine phosphatase RsbU (Q94L) dimer, 77 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 24
|
A general mechanism for initiating the bacterial general stress response.
Elife 13 (2025)
Baral R, Ho K, Kumar RP, Hopkins JB, Watkins MB, LaRussa S, Caban-Penix S, Calderone LA, Bradshaw N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
119 |
nm3 |
|
|
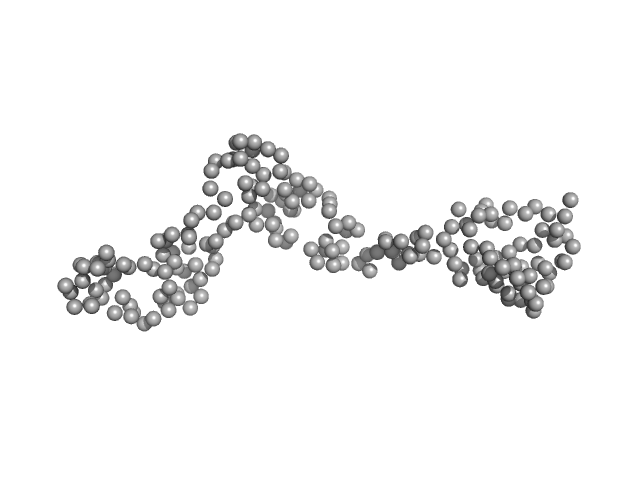
UniProt ID: Q9Z1X4 (395-592) Interleukin enhancer-binding factor 3
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 21 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: Q9Z1X4 (1-381) Interleukin enhancer-binding factor 3
UniProt ID: Q9CXY6 (29-390) Interleukin enhancer-binding factor 2
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 42 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
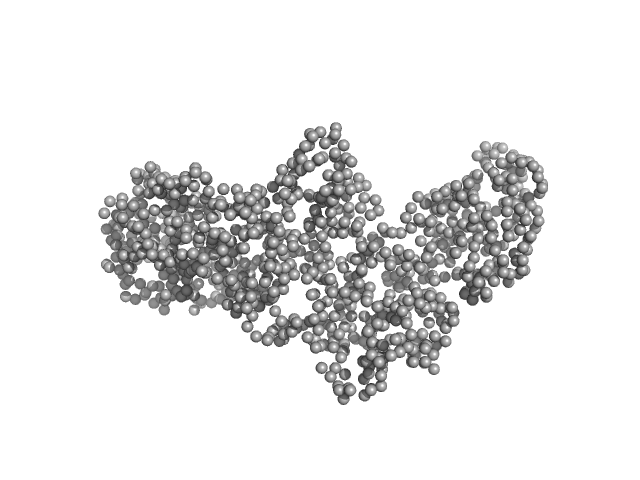
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: None (None-None) 25-mer dsRNA
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
25-mer dsRNA monomer, 16 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.9 |
nm |
| VolumePorod |
453 |
nm3 |
|
|
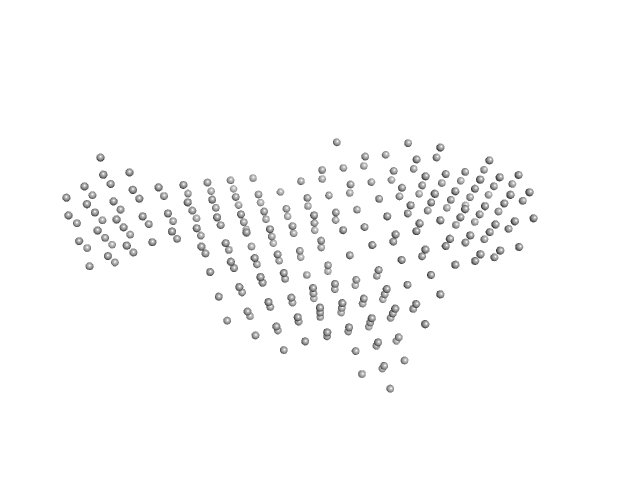
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: None (None-None) 25-mer dsRNA
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
25-mer dsRNA monomer, 16 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
431 |
nm3 |
|
|
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: None (None-None) 36-mer dsRNA
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
36-mer dsRNA monomer, 23 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
587 |
nm3 |
|
|