UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: None (None-None) 36-mer dsRNA
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
36-mer dsRNA monomer, 23 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.1 |
nm |
| VolumePorod |
626 |
nm3 |
|
|
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: None (None-None) 54-mer dsRNA
|
|
|
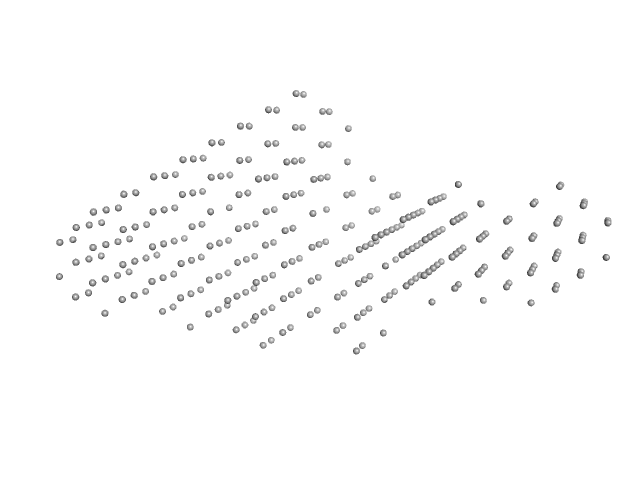
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.1 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
797 |
nm3 |
|
|
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: None (None-None) 54-mer dsRNA
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
|
|
|
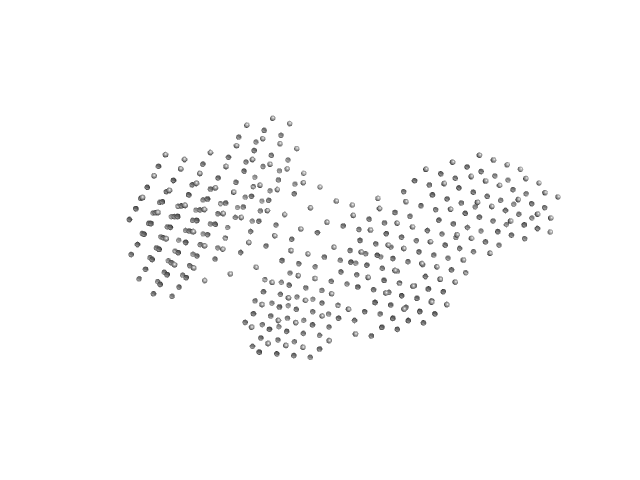
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.7 |
nm |
| Dmax |
26.5 |
nm |
| VolumePorod |
1075 |
nm3 |
|
|
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: None (None-None) 54-mer dsRNA
UniProt ID: Q9Z1X4 (2-591) Interleukin enhancer-binding factor 3
UniProt ID: Q12905 (1-390) Interleukin enhancer-binding factor 2
|
|
|
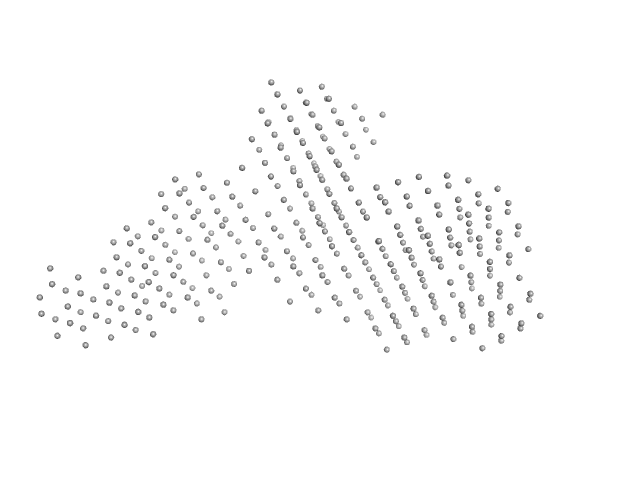
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.8 |
nm |
| Dmax |
26.1 |
nm |
| VolumePorod |
1050 |
nm3 |
|
|
UniProt ID: A0QSU3 (2-513) Inosine-5'-monophosphate dehydrogenase
|
|
|
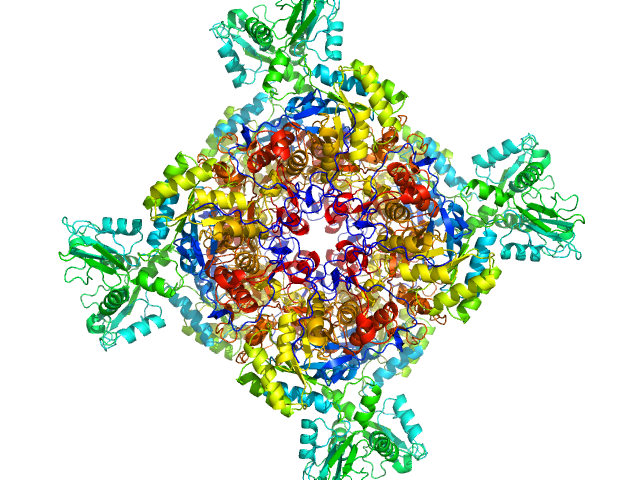
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Feb 21
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.3 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
952 |
nm3 |
|
|
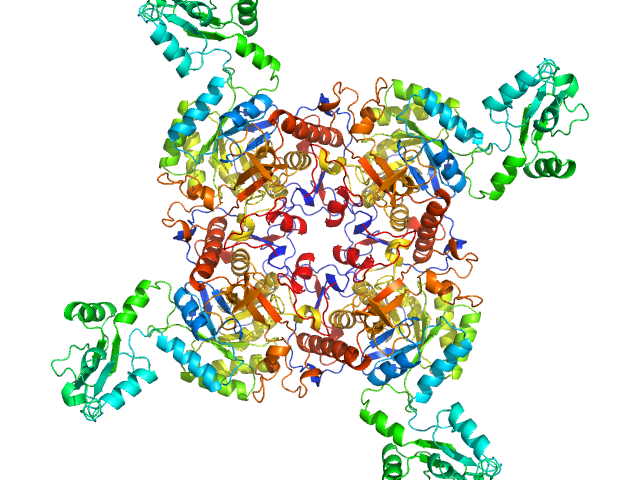
UniProt ID: A0QSU3 (2-513) Inosine-5'-monophosphate dehydrogenase
|
|
|
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase tetramer, 213 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Mar 6
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
471 |
nm3 |
|
|
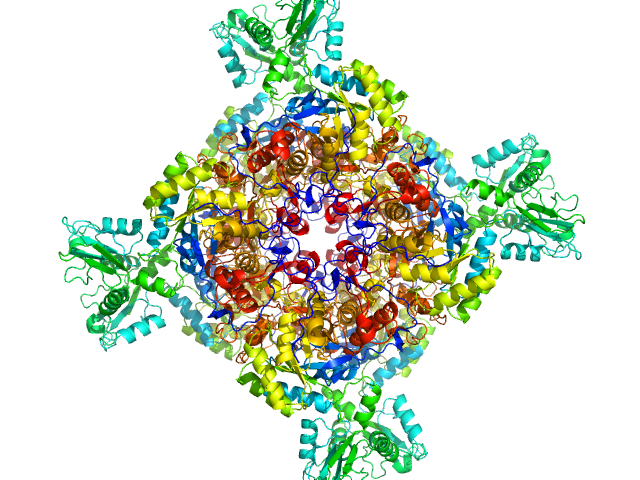
UniProt ID: A0QSU3 (2-513) Inosine-5'-monophosphate dehydrogenase
|
|
|
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Jan 29
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
821 |
nm3 |
|
|
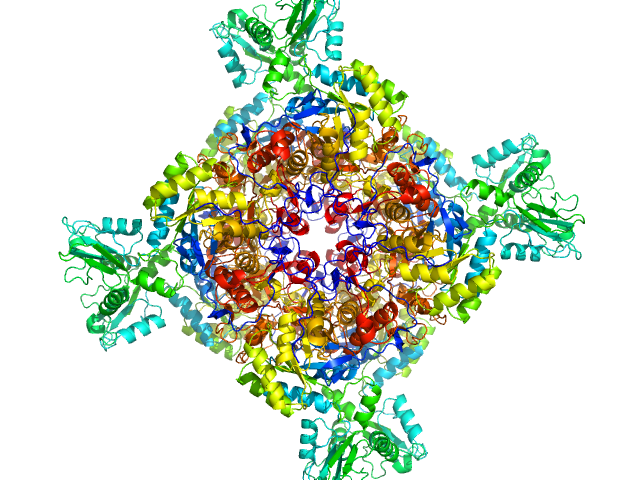
UniProt ID: A0QSU3 (2-513) Inosine-5'-monophosphate dehydrogenase
|
|
|
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Mar 6
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.1 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
784 |
nm3 |
|
|
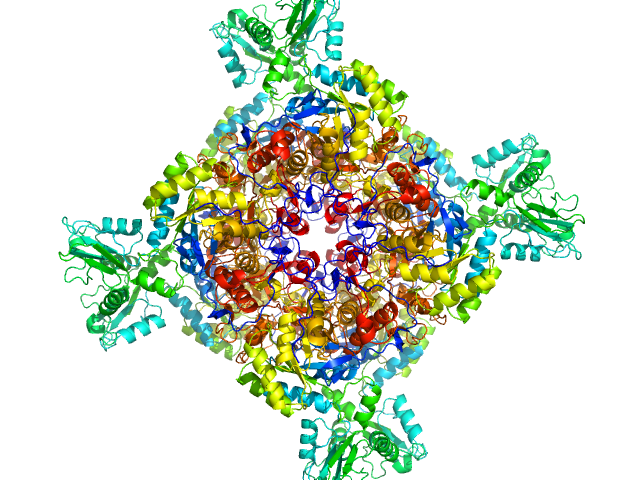
UniProt ID: A0QSU3 (2-513) Inosine-5'-monophosphate dehydrogenase
|
|
|
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Mar 6
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
810 |
nm3 |
|
|
UniProt ID: P61825 (756-836) Structural polyprotein
|
|
|
|
| Sample: |
Structural polyprotein dimer, 26 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 27
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|