UniProt ID: Q6D8U3 (26-924) Ferredoxin protease E83A mutant
UniProt ID: P16972 (53-145) Arabidopsis ferredoxin 2
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
UniProt ID: Q6D8U3 (26-924) Ferredoxin protease E83A mutant
UniProt ID: P16972 (53-145) Arabidopsis ferredoxin 2
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
178 |
nm3 |
|
|
UniProt ID: Q6D8U3 (26-924) Ferredoxin protease E83A mutant
UniProt ID: P16972 (53-145) Arabidopsis ferredoxin 2
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
UniProt ID: Q6D8U3 (26-924) Ferredoxin protease E83A mutant
UniProt ID: P16972 (53-145) Arabidopsis ferredoxin 2
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
UniProt ID: P02649 (19-317) Apolipoprotein E2
|
|
|
|
| Sample: |
Apolipoprotein E2 tetramer, 139 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
The molecular basis for Apolipoprotein E4 as the major risk factor for late onset Alzheimer's disease.
J Mol Biol (2019)
Raulin AC, Kraft L, Al-Hilaly YK, Xue WF, McGeehan JE, Atack JR, Serpell L
|
| RgGuinier |
5.6 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
UniProt ID: Q9NRI5 (691-836) Disrupted- in-schizophrenia 1 (DISC1 12D2) 691-836
|
|
|
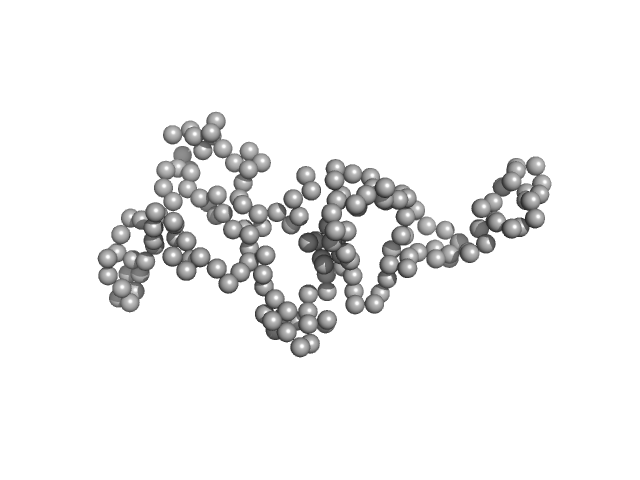
|
| Sample: |
Disrupted- in-schizophrenia 1 (DISC1 12D2) 691-836 monomer, 19 kDa protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 1
|
Biophysical insights from a single chain camelid antibody directed against the Disrupted-in-Schizophrenia 1 protein.
PLoS One 13(1):e0191162 (2018)
Yerabham ASK, Müller-Schiffmann A, Ziehm T, Stadler A, Köber S, Indurkhya X, Marreiros R, Trossbach SV, Bradshaw NJ, Prikulis I, Willbold D, Weiergräber OH, Korth C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: None (None-None) anti-DISC1 single-domain camelid antibody VHH B5
UniProt ID: Q9NRI5 (691-836) Disrupted- in-schizophrenia 1 (DISC1 12D2) 691-836
|
|
|

|
| Sample: |
Anti-DISC1 single-domain camelid antibody VHH B5 monomer, 14 kDa protein
Disrupted- in-schizophrenia 1 (DISC1 12D2) 691-836 monomer, 19 kDa protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 19
|
Biophysical insights from a single chain camelid antibody directed against the Disrupted-in-Schizophrenia 1 protein.
PLoS One 13(1):e0191162 (2018)
Yerabham ASK, Müller-Schiffmann A, Ziehm T, Stadler A, Köber S, Indurkhya X, Marreiros R, Trossbach SV, Bradshaw NJ, Prikulis I, Willbold D, Weiergräber OH, Korth C
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E248A/E251A C1
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E248A/E251A C1 dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, and 5% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 May 26
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E248A/E251A C1
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E248A/E251A C1 dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, and 5% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 May 26
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E248A/E251A C1
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E248A/E251A C1 dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, and 5% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 May 26
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
95 |
nm3 |
|
|