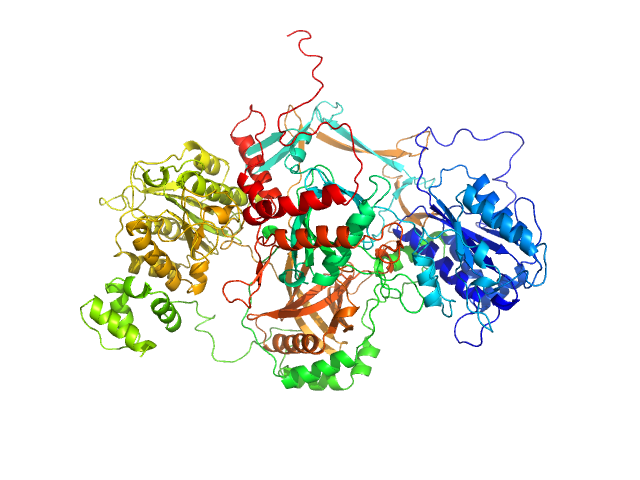
UniProt ID: P12956 (1-609) X-ray repair cross-complementing protein 6
UniProt ID: P13010 (1-565) X-ray repair cross-complementing protein 5 ΔCTR
|
|
|
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 ΔCTR monomer, 64 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
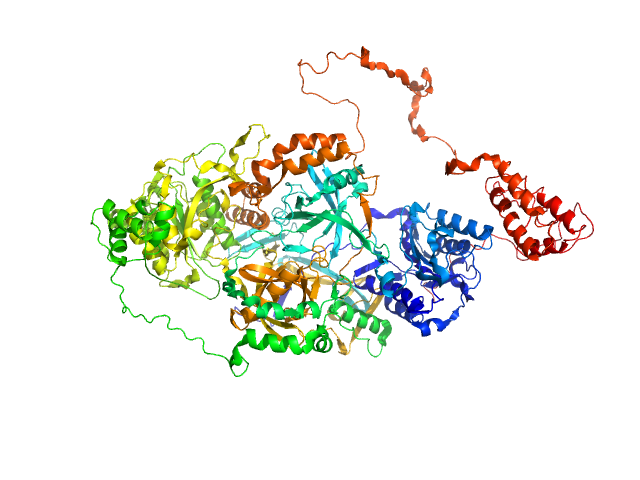
UniProt ID: P12956 (1-609) X-ray repair cross-complementing protein 6
UniProt ID: P13010 (1-732) X-ray repair cross-complementing protein 5
|
|
|
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
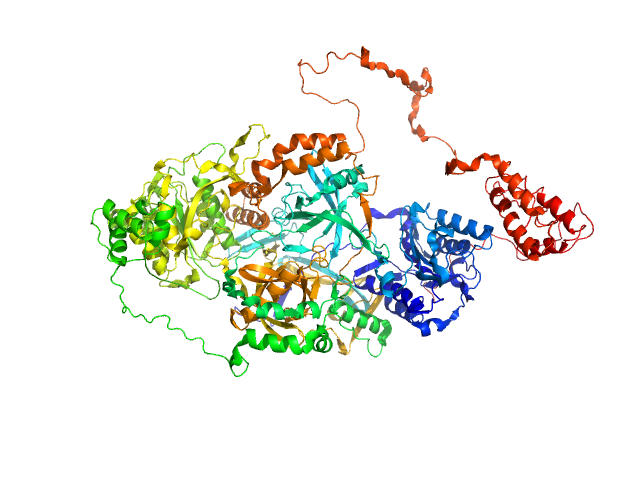
UniProt ID: P12956 (1-609) X-ray repair cross-complementing protein 6
UniProt ID: P13010 (1-732) X-ray repair cross-complementing protein 5
UniProt ID: None (None-None) Y-DNA
|
|
|
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
Y-DNA monomer, 18 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Jan 8
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
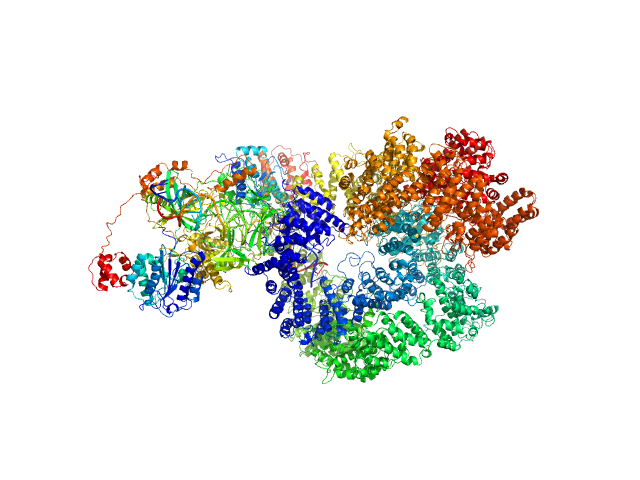
UniProt ID: P12956 (1-609) X-ray repair cross-complementing protein 6
UniProt ID: P13010 (1-732) X-ray repair cross-complementing protein 5
UniProt ID: P78527 (10-4128) DNA-dependent protein kinase catalytic subunit
UniProt ID: None (None-None) dsDNA
|
|
|
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
6.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
1090 |
nm3 |
|
|
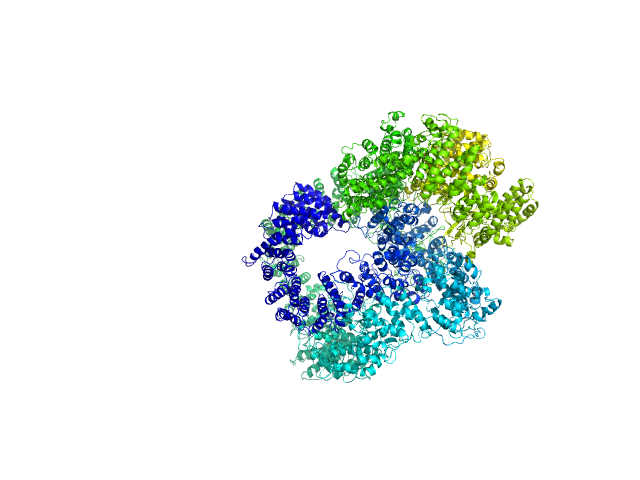
UniProt ID: P12956 (1-609) X-ray repair cross-complementing protein 6
UniProt ID: P13010 (1-732) X-ray repair cross-complementing protein 5
UniProt ID: P78527 (10-4128) DNA-dependent protein kinase catalytic subunit
UniProt ID: None (None-None) dsDNA
|
|
|
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
7.5 |
nm |
| Dmax |
29.4 |
nm |
| VolumePorod |
1440 |
nm3 |
|
|