|
|
|
|
|
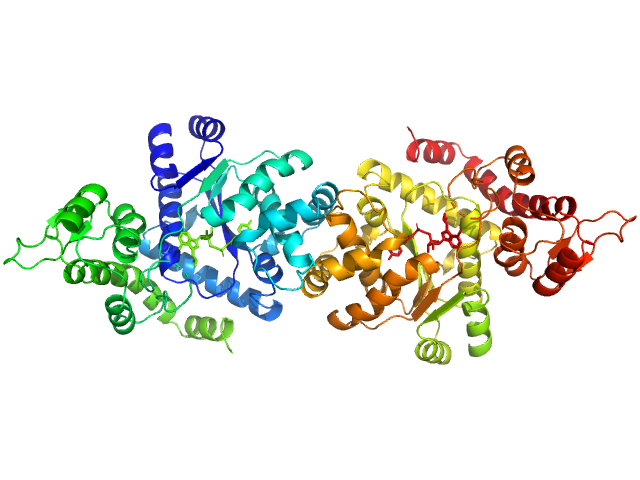
| Sample: |
Tyrosine--tRNA ligase, cytoplasmic dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4
|
| Experiment: |
SAXS
data collected at ...WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D...
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
118 |
nm3 |
|