|
|
|
|
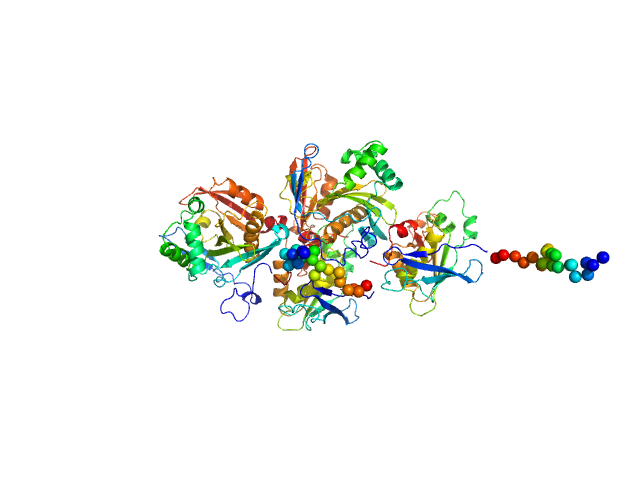
|
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF (Δ444-462) dimer, 107 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2023 Jan 18
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
559 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.0 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
546 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
563 |
nm3 |
|
|
|
|
|
|
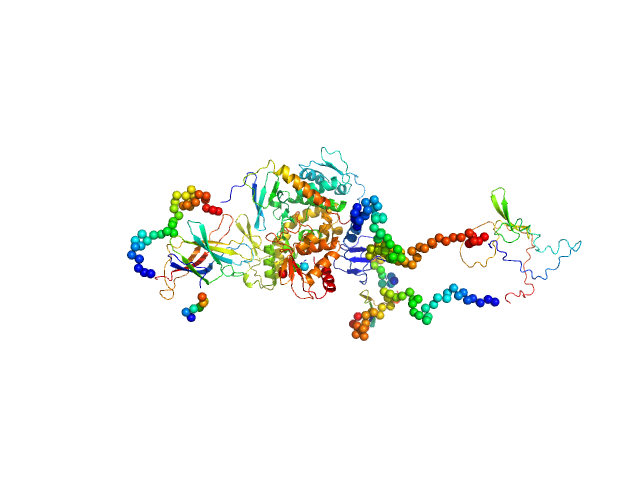
|
| Sample: |
Isoform 5 of E3 ubiquitin-protein ligase NEDD4-like monomer, 110 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 1 mM EDTA, 3% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Aug 17
|
Structural basis of ubiquitin ligase Nedd4-2 autoinhibition and regulation by calcium and 14-3-3 proteins
Nature Communications 16(1) (2025)
Janosev M, Kosek D, Tekel A, Joshi R, Honzejkova K, Pohl P, Obsil T, Obsilova V
|
| RgGuinier |
4.9 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform 5 of E3 ubiquitin-protein ligase NEDD4-like monomer, 110 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 1 mM CaCl2, 3% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Aug 17
|
Structural basis of ubiquitin ligase Nedd4-2 autoinhibition and regulation by calcium and 14-3-3 proteins
Nature Communications 16(1) (2025)
Janosev M, Kosek D, Tekel A, Joshi R, Honzejkova K, Pohl P, Obsil T, Obsilova V
|
| RgGuinier |
4.9 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
|
|
|
|
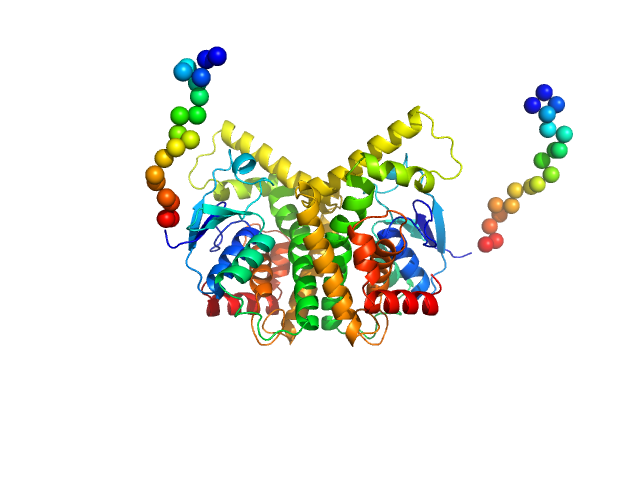
|
| Sample: |
GST N-terminal domain-containing protein dimer, 60 kDa Gordonia rubripertincta protein
|
| Buffer: |
10 mM Tris, 300 mM NaCl,, pH: 8
|
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2022 Jan 20
|
Glutathione S-Transferase Mediated Epoxide Conversion: Structural Insights into an Enantioselective Catalyst
Jens Reiners
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aminoacyl tRNA synthase complex-interacting multifunctional protein 1 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2022 Oct 12
|
Solution structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex
Michal Taube
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C3H1-type domain-containing protein monomer, 8 kDa Selaginella moellendorffii protein
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 9
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C3H1-type domain-containing protein monomer, 8 kDa Selaginella moellendorffii protein
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 9
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
14 |
nm3 |
|
|