|
|
|
|
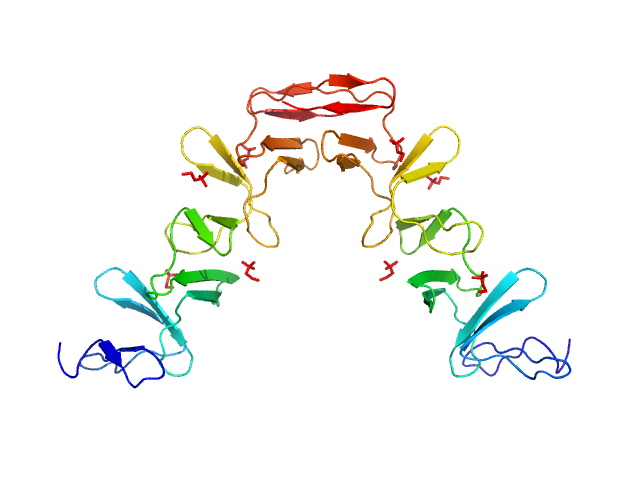
|
| Sample: |
Lytic Amidase choline-binding domain dimer, 17 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 11
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Functional binding region (187-385) of the pneumococcal serine-rich repeat protein monomer, 22 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM sodium citrate 250 mM NaCl 2.5 % Glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
The basic keratin 10-binding domain of the virulence-associated pneumococcal serine-rich protein PsrP adopts a novel MSCRAMM fold.
Open Biol 4:130090 (2014)
Schulte T, Löfling J, Mikaelsson C, Kikhney A, Hentrich K, Diamante A, Ebel C, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2009 Nov 19
|
The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping.
Proc Natl Acad Sci U S A 111(1):457-62 (2014)
Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
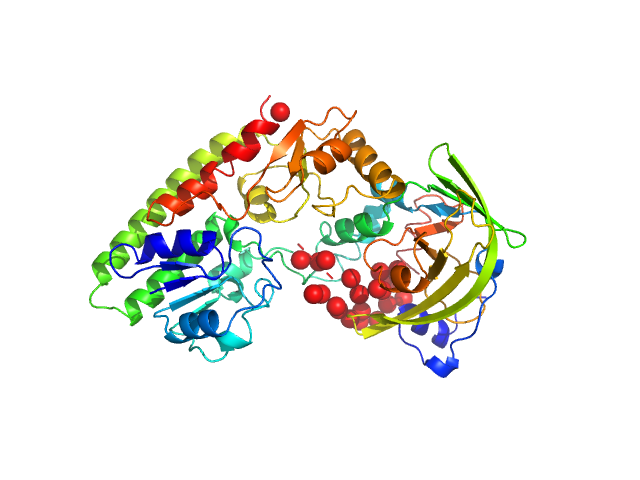
|
| Sample: |
High-affinity zinc transporter periplasmic component monomer, 33 kDa Salmonella enterica subsp. … protein
Zinc/cadmium-binding protein monomer, 23 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 May 9
|
The Salmonella enterica ZinT structure, zinc affinity and interaction with the high-affinity uptake protein ZnuA provide insight into the management of periplasmic zinc.
Biochim Biophys Acta 1840(1):535-44 (2014)
Ilari A, Alaleona F, Tria G, Petrarca P, Battistoni A, Zamparelli C, Verzili D, Falconi M, Chiancone E
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
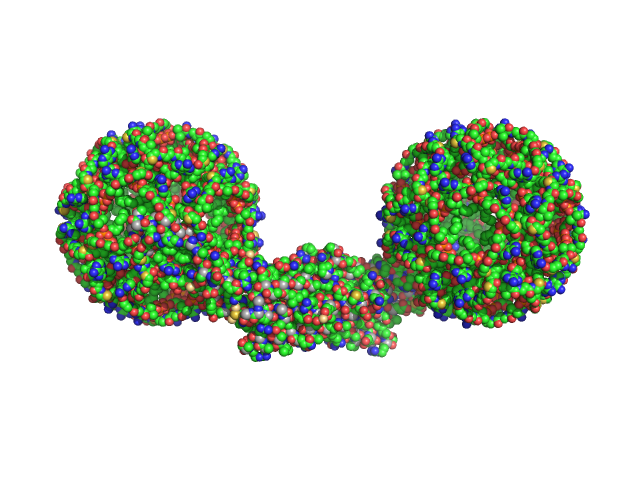
|
| Sample: |
Endophilin-A1 BAR domain dimer, 58 kDa Mus musculus protein
Arachidonyl-CoA, 60 kDa
|
| Buffer: |
50 mM TRIS-HCL 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 27
|
Endophilin-A1 BAR domain interaction with arachidonyl CoA.
Front Mol Biosci 1:20 (2014)
Petoukhov MV, Weissenhorn W, Svergun DI
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
480 |
nm3 |
|
|
|
|
|
|
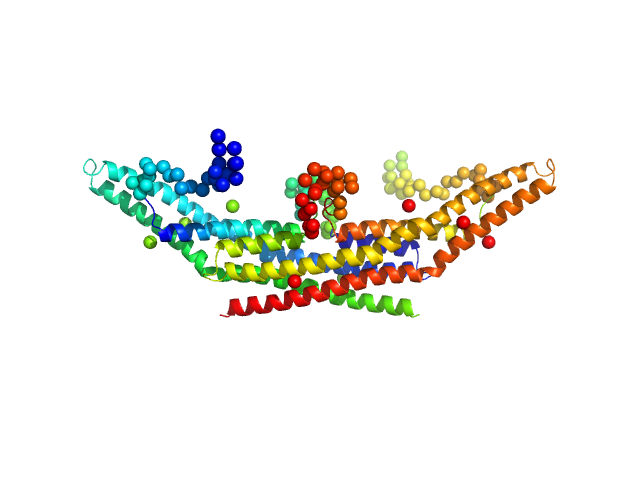
|
| Sample: |
Endophilin-A1 BAR domain dimer, 58 kDa Mus musculus protein
|
| Buffer: |
50 mM TRIS-HCL 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 13
|
Endophilin-A1 BAR domain interaction with arachidonyl CoA.
Front Mol Biosci 1:20 (2014)
Petoukhov MV, Weissenhorn W, Svergun DI
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Modification methylase SsoII monomer, 43 kDa Shigella sonnei protein
12-bp DNA monomer, 8 kDa DNA
|
| Buffer: |
50 mM Na-phosphate buffer, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Mar 13
|
Flexibility of the linker between the domains of DNA methyltransferase SsoII revealed by small-angle X-ray scattering: implications for transcription regulation in SsoII restriction-modification system.
PLoS One 9(4):e93453 (2014)
Konarev PV, Kachalova GS, Ryazanova AY, Kubareva EA, Karyagina AS, Bartunik HD, Svergun DI
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
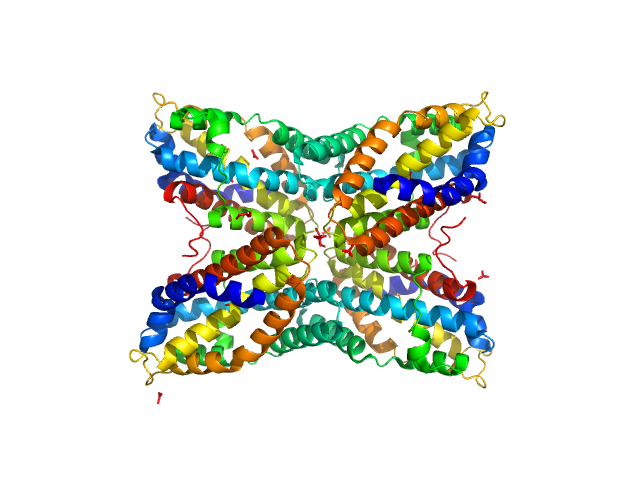
| Sample: |
Thiaminase type II enzyme tetramer, 107 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 11
|
Staphylococcus aureus thiaminase II: oligomerization warrants proteolytic protection against serine proteases.
Acta Crystallogr D Biol Crystallogr 69(Pt 12):2320-9 (2013)
Begum A, Drebes J, Kikhney A, Müller IB, Perbandt M, Svergun D, Wrenger C, Betzel C
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - 5α-cholestan-3-one) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.1 |
nm |
| Dmax |
20.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - 5α-cholestan-3-one) with Dox (10%) monomer, 220 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
7.5 |
nm |
| Dmax |
25.5 |
nm |
|
|