|
|
|
|
|
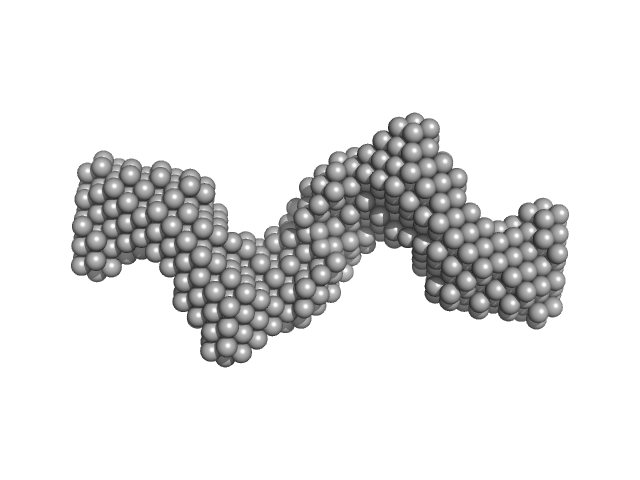
| Sample: |
Clostridium difficile bacteriophage 27 endolysin C238R mutant dimer, 64 kDa Clostridioides difficile protein
|
| Buffer: |
20 mM HEPES 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 21
|
The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.
PLoS Pathog 10(7):e1004228 (2014)
Dunne M, Mertens HD, Garefalaki V, Jeffries CM, Thompson A, Lemke EA, Svergun DI, Mayer MJ, Narbad A, Meijers R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ataxin-3 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
6.9 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
425 |
nm3 |
|
|
|
|
|
|
|
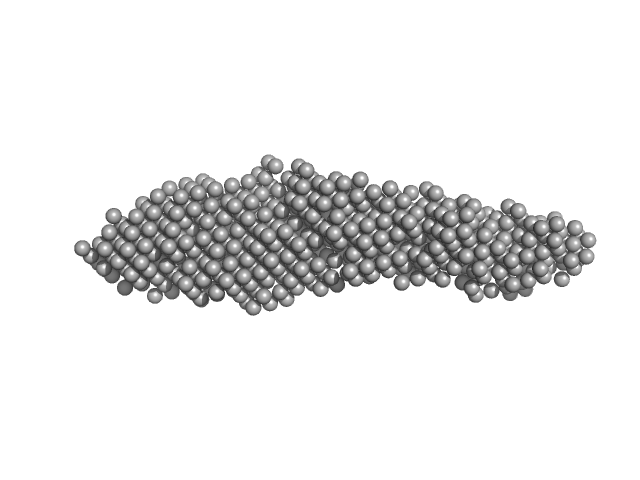
| Sample: |
Tubulin alpha-1A chain monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
7.0 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ataxin-3 monomer, 41 kDa Homo sapiens protein
Tubulin alpha-1A chain monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
8.4 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
900 |
nm3 |
|
|
|
|
|
|
|
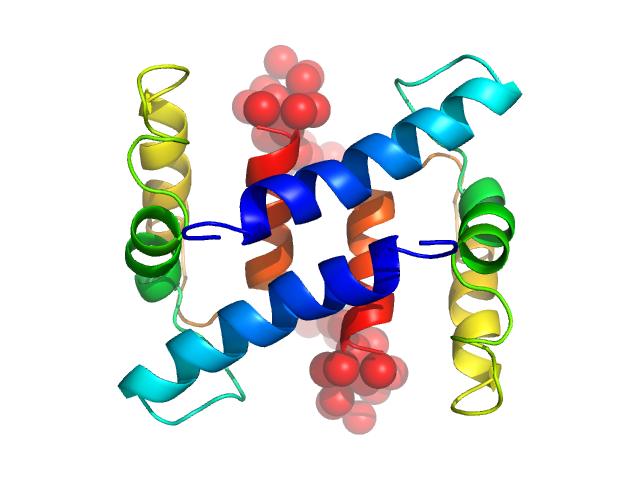
| Sample: |
Protein S100-A4 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 20 mM NaCl, 0.1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Mar 2
|
The C-Terminal Random Coil Region Tunes the Ca2+-Binding Affinity of S100A4 through Conformational Activation
PLoS ONE 9(5):e97654 (2014)
Duelli A, Kiss B, Lundholm I, Bodor A, Petoukhov M, Svergun D, Nyitray L, Katona G, Csernoch L
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myelin basic protein monomer, 18 kDa Bos taurus protein
|
| Buffer: |
20 mM NaH2PO4/ Na2HPO4, 99.9% D2O, pH: 4.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Feb 21
|
Internal nanosecond dynamics in the intrinsically disordered myelin basic protein.
J Am Chem Soc 136(19):6987-94 (2014)
Stadler AM, Stingaciu L, Radulescu A, Holderer O, Monkenbusch M, Biehl R, Richter D
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.1 |
nm |
|
|
|
|
|
|
|
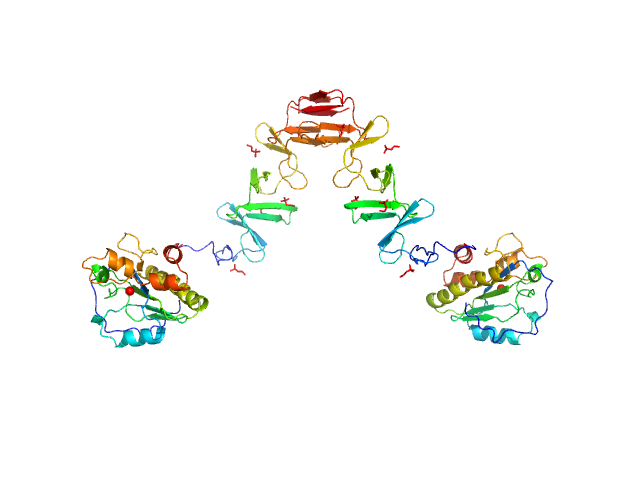
| Sample: |
Chitinase 60 monomer, 61 kDa Moritella marina protein
|
| Buffer: |
20 mM Tris 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 18
|
Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution.
Acta Crystallogr D Biol Crystallogr 70(Pt 3):676-84 (2014)
Malecki PH, Vorgias CE, Petoukhov MV, Svergun DI, Rypniewski W
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
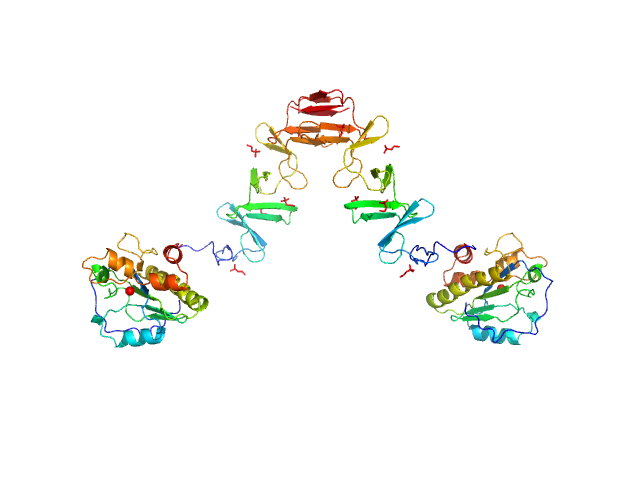
| Sample: |
Lytic Amidase with choline dimer, 75 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
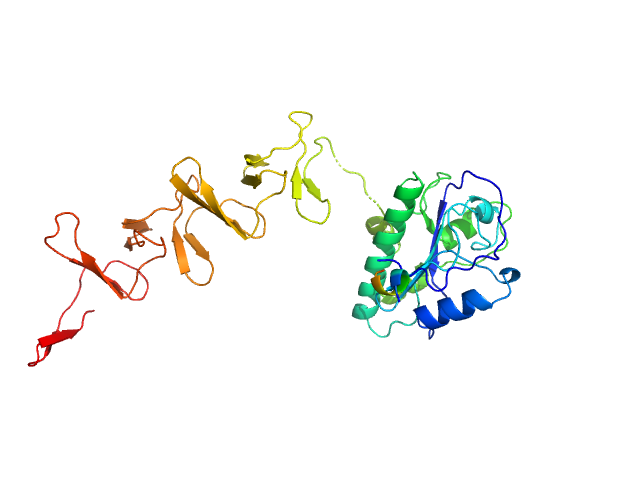
| Sample: |
Prophage Lytic Amidase with choline dimer, 73 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
5.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lytic Amidase without choline monomer, 37 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 3
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|