|
|
|
|
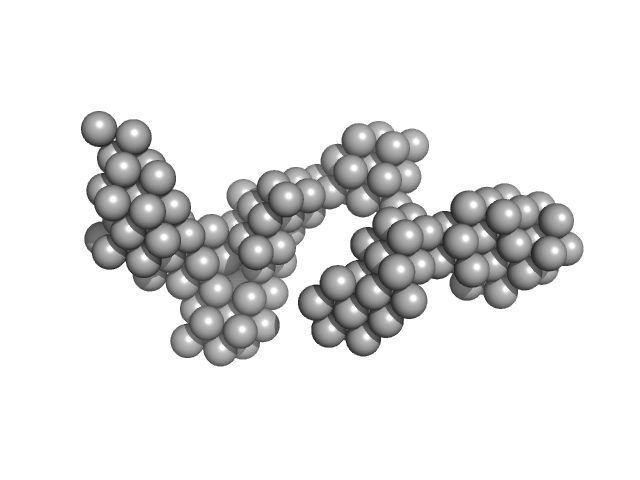
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Opb-Chol) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.5 |
nm |
|
|
|
|
|
|
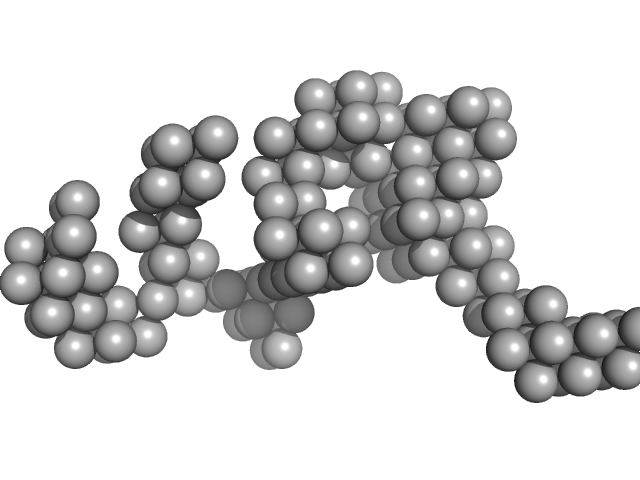
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Opb-Chol) with Dox (10%) monomer, 225 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
10.0 |
nm |
| Dmax |
21.9 |
nm |
|
|
|
|
|
|
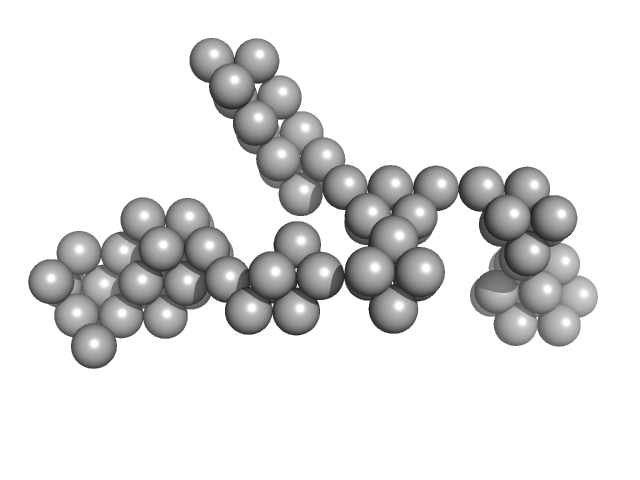
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Lev-Chol) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.5 |
nm |
| Dmax |
22.9 |
nm |
|
|
|
|
|
|
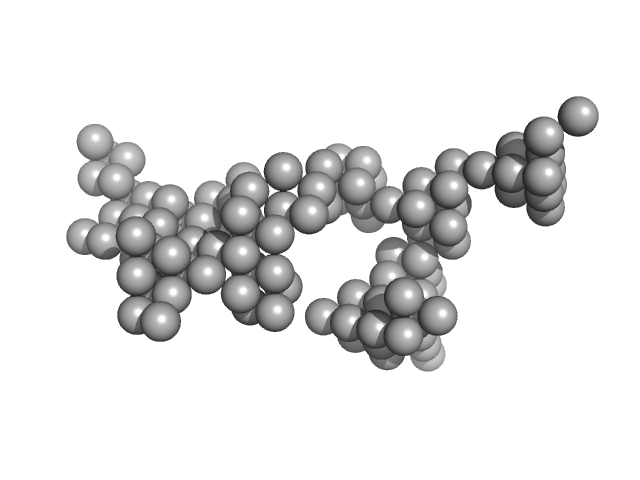
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Lev-Chol) with Dox (10%) monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.9 |
nm |
| Dmax |
23.8 |
nm |
|
|
|
|
|
|
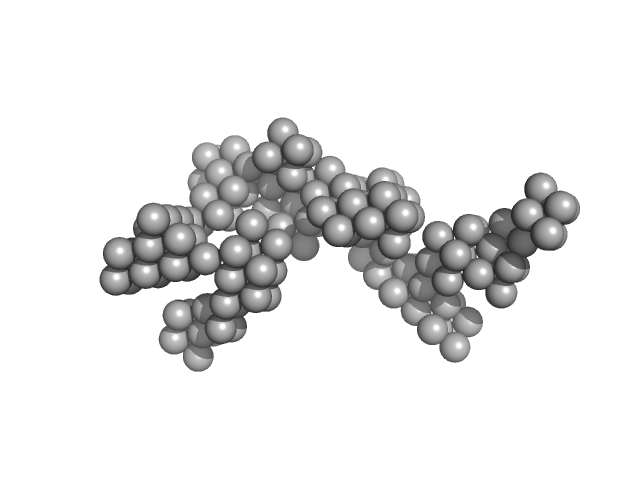
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - cholest-4-en-3-one) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 16
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
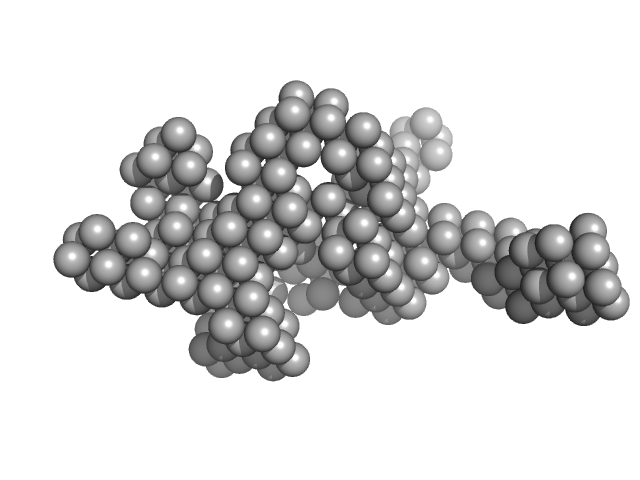
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - cholest-4-en-3-one) with Dox (10%) monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
5.5 |
nm |
| Dmax |
18.3 |
nm |
|
|
|
|
|
|
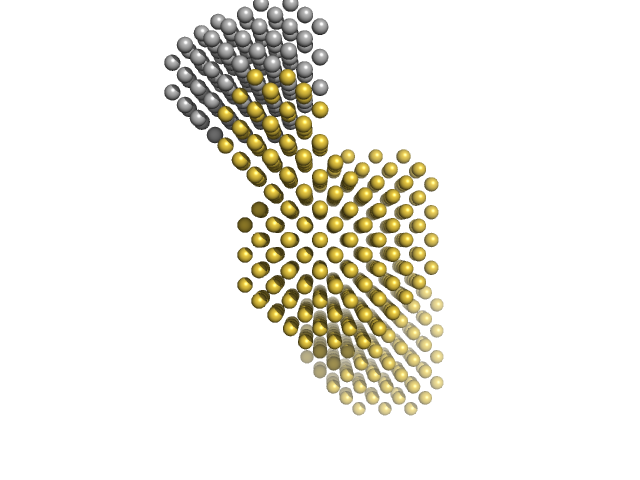
|
| Sample: |
I27-PimA Fusion protein monomer, 48 kDa protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 May 17
|
Conformational plasticity of the essential membrane-associated mannosyltransferase PimA from mycobacteria.
J Biol Chem 288(41):29797-808 (2013)
Giganti D, Alegre-Cebollada J, Urresti S, Albesa-Jové D, Rodrigo-Unzueta A, Comino N, Kachala M, López-Fernández S, Svergun DI, Fernández JM, Guerin ME
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.8 |
nm |
|
|