|
|
|
|

|
| Sample: |
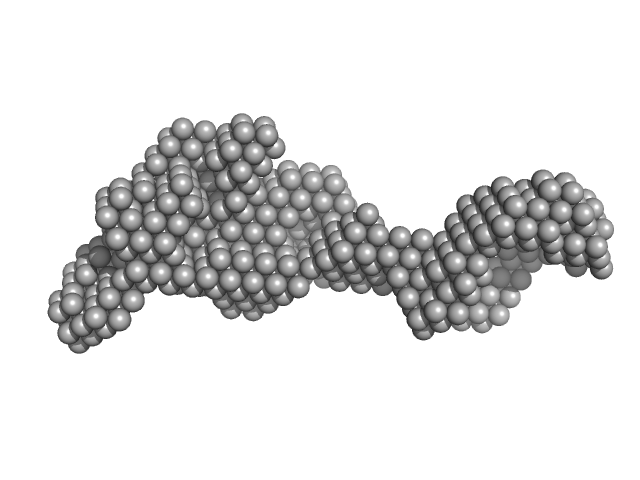
DNA mismatch repair protein MutS dimer, 191 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Feb 28
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
307 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
7.8 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
700 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
8.5 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
750 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
8.3 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
720 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
7.8 |
nm |
| Dmax |
27.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
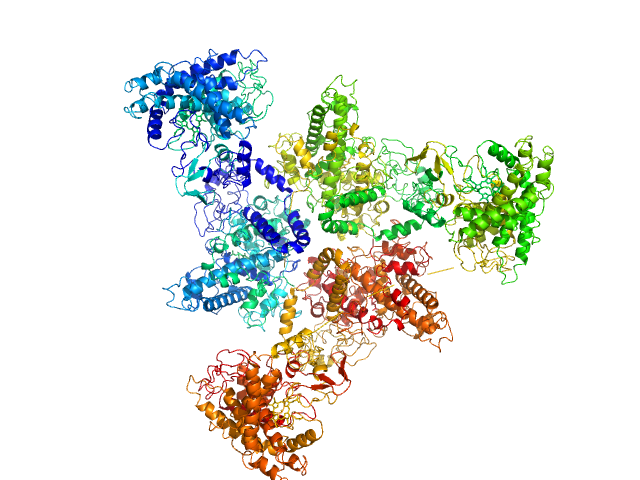
Psi-producing oxygenase A trimer, 362 kDa Aspergillus nidulans protein
|
| Buffer: |
20 Mm HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 24
|
A structural model of PpoA derived from SAXS-analysis-implications for substrate conversion.
Biochim Biophys Acta 1831(9):1449-57 (2013)
Koch C, Tria G, Fielding AJ, Brodhun F, Valerius O, Feussner K, Braus GH, Svergun DI, Bennati M, Feussner I
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
540 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
GTP-binding nuclear protein Ran monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
GTP-binding nuclear protein Ran monomer, 24 kDa Homo sapiens protein
Snurportin-1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
|
|