|
|
|
|
|
| Sample: |
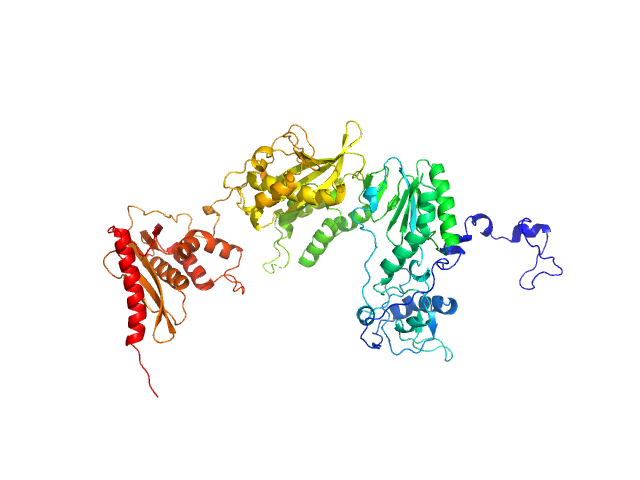
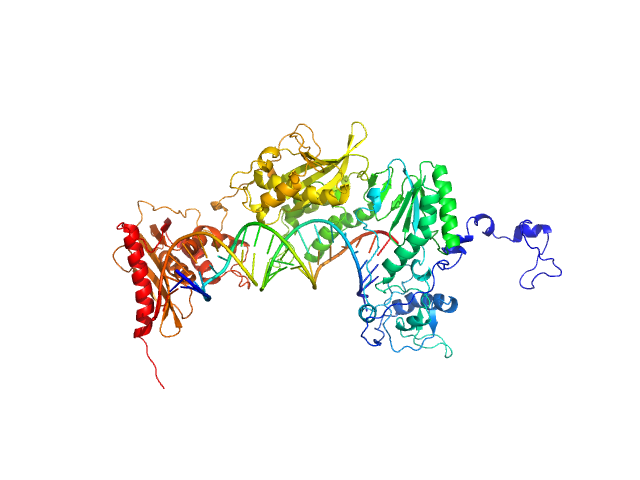
Exportin-1 monomer, 123 kDa Mus musculus protein
Snurportin-1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 10
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDLV4_fit1_model1.png)
|
| Sample: |
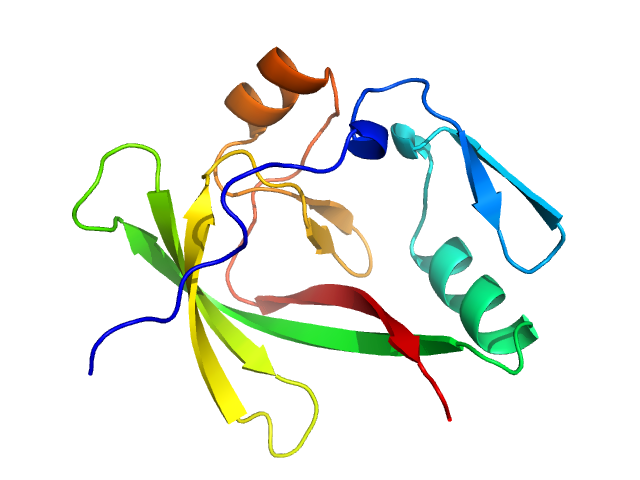
Ceruloplasmin dimer, 244 kDa Homo sapiens protein
Lactotransferrin dimer, 156 kDa Homo sapiens protein
Myeloperoxidase dimer, 132 kDa Homo sapiens protein
|
| Buffer: |
0.1 M sodium-acetate buffer, pH: 5.6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Nov 24
|
Ceruloplasmin: Macromolecular Assemblies with Iron-Containing Acute Phase Proteins
PLoS ONE 8(7):e67145 (2013)
Samygina V, Sokolov A, Bourenkov G, Petoukhov M, Pulina M, Zakharova E, Vasilyev V, Bartunik H, Svergun D, Tuma R
|
| RgGuinier |
6.9 |
nm |
| Dmax |
22.6 |
nm |
| VolumePorod |
726 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apo XMRV RT monomer, 75 kDa Escherichia coli protein
|
| Buffer: |
10 mM HEPES 100 mM KCl 5% Glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 8
|
Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid.
Nucleic Acids Res 41(6):3874-87 (2013)
Nowak E, Potrzebowski W, Konarev PV, Rausch JW, Bona MK, Svergun DI, Bujnicki JM, Le Grice SF, Nowotny M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apo XMRV RT monomer, 75 kDa Escherichia coli protein
RNA_DNA hybrid substrate monomer, 15 kDa
|
| Buffer: |
10 mM HEPES 100 mM KCl 5% Glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 8
|
Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid.
Nucleic Acids Res 41(6):3874-87 (2013)
Nowak E, Potrzebowski W, Konarev PV, Rausch JW, Bona MK, Svergun DI, Bujnicki JM, Le Grice SF, Nowotny M
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ataxin-1 monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 21
|
Self-assembly and conformational heterogeneity of the AXH domain of ataxin-1: an unusual example of a chameleon fold.
Biophys J 104(6):1304-13 (2013)
de Chiara C, Rees M, Menon RP, Pauwels K, Lawrence C, Konarev PV, Svergun DI, Martin SR, Chen YW, Pastore A
|
|
|
|
|
|
|
|
| Sample: |
Peroxisomal multifunctional enzyme type 2 dimer, 128 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Sodium Phosphate 200 mM NaCl 5% (v/v) Glycerol 1mM Na2EDTA 1 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 12
|
Quaternary structure of human, Drosophila melanogaster and Caenorhabditis elegans MFE-2 in solution from synchrotron small-angle X-ray scattering.
FEBS Lett 587(4):305-10 (2013)
Mehtälä ML, Haataja TJ, Blanchet CE, Hiltunen JK, Svergun DI, Glumoff T
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Peroxisomal multifunctional enzyme type 2 dimer, 159 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 200 mM NaCl 5% (v/v) Glycerol 1mM Na2EDTA 1 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 22
|
Quaternary structure of human, Drosophila melanogaster and Caenorhabditis elegans MFE-2 in solution from synchrotron small-angle X-ray scattering.
FEBS Lett 587(4):305-10 (2013)
Mehtälä ML, Haataja TJ, Blanchet CE, Hiltunen JK, Svergun DI, Glumoff T
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Polcalcin Phl p 7 monomer, 9 kDa Phleum pratense protein
|
| Buffer: |
0.15M NaCl, 0.025M Hepes, pH 7.4, 100 uM Ca2+, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Dec 8
|
Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg2+-bound, and fully Ca2+-bound.
Proteins 81(2):300-15 (2013)
Henzl MT, Sirianni AG, Wycoff WG, Tan A, Tanner JJ
|
| RgGuinier |
1.3 |
nm |
| Dmax |
3.6 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Activator of Hsp90 ATPase-1 monomer, 38 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Sodium phosphate, 50 mM NaCl, 2 mM EDTA, 1 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2013 Jun 1
|
Low resolution structural studies indicate that the activator of Hsp90 ATPase 1 (Aha1) of Leishmania braziliensis has an elongated shape which allows its interaction with both N- and M-domains of Hsp90.
PLoS One 8(6):e66822 (2013)
Seraphim TV, Alves MM, Silva IM, Gomes FE, Silva KP, Murta SM, Barbosa LR, Borges JC
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Adaptor protein, phosphotyrosine interaction, pleckstrin homology domain, and leucine zipper-containing protein 2 dimer, 87 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 7 Apr 20
|
Membrane curvature protein exhibits interdomain flexibility and binds a small GTPase.
J Biol Chem 287(49):40996-1006 (2012)
King GJ, Stöckli J, Hu SH, Winnen B, Duprez WG, Meoli CC, Junutula JR, Jarrott RJ, James DE, Whitten AE, Martin JL
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|