|
|
|
|

|
| Sample: |
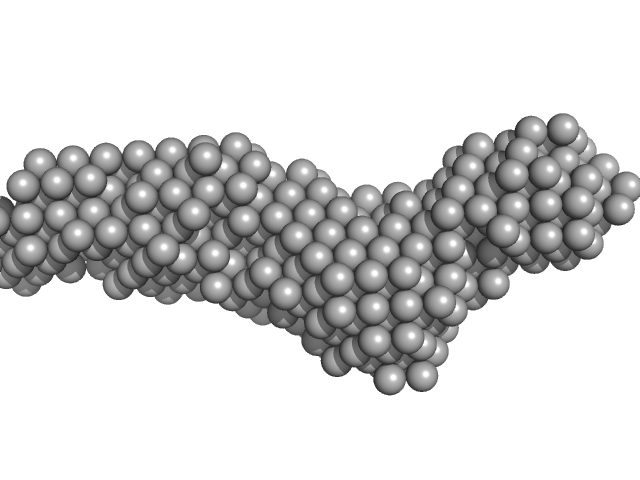
Collagenase ColG segement s2s3as3b monomer, 37 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
N-terminus of disulfide interchange protein DsbD monomer, 14 kDa Neisseria meningitidis protein
|
| Buffer: |
25mM HEPES 150mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Production, biophysical characterization and initial crystallization studies of the N- and C-terminal domains of DsbD, an essential enzyme in Neisseria meningitidis.
Acta Crystallogr F Struct Biol Commun 74(Pt 1):31-38 (2018)
Smith RP, Whitten AE, Paxman JJ, Kahler CM, Scanlon MJ, Heras B
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein) monomer, 21 kDa Wolbachia endosymbiont of … protein
|
| Buffer: |
25mM HEPES 150mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Production, biophysical characterization and initial crystallization studies of the N- and C-terminal domains of DsbD, an essential enzyme in Neisseria meningitidis.
Acta Crystallogr F Struct Biol Commun 74(Pt 1):31-38 (2018)
Smith RP, Whitten AE, Paxman JJ, Kahler CM, Scanlon MJ, Heras B
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColG segement s2s3as3b monomer, 37 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 11
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human respiratory syncytial virus M2-1 monomer, 14 kDa Human orthopneumovirus protein
|
| Buffer: |
20 mM Tris–HCl, 300 mM NaCl,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 13
|
Structure and stability of the Human respiratory syncytial virus M2-1 RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun 74(Pt 1):23-30 (2018)
Molina IG, Josts I, Almeida Hernandez Y, Esperante S, Salgueiro M, Garcia Alai MM, de Prat-Gay G, Tidow H
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
NHL repeat-containing protein 2 monomer, 80 kDa Homo sapiens protein
|
| Buffer: |
20 mM BisTris, 100 mM NaCl, 2 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jul 8
|
Structural analysis of human NHLRC2, mutations of which are associated with FINCA disease.
PLoS One 13(8):e0202391 (2018)
Biterova E, Ignatyev A, Uusimaa J, Hinttala R, Ruddock LW
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
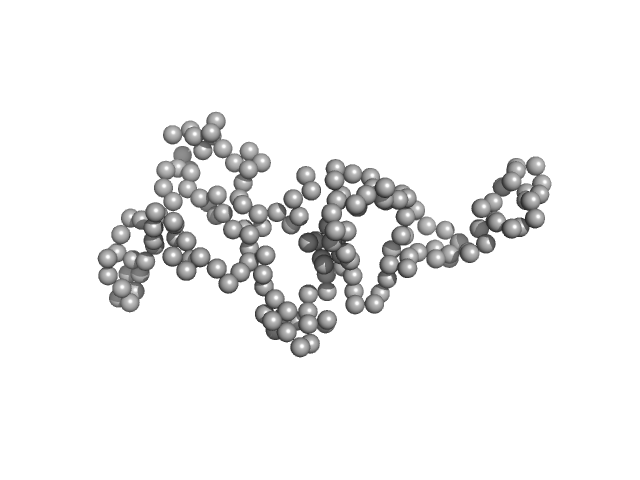
|
| Sample: |
Disrupted- in-schizophrenia 1 (DISC1 12D2) 691-836 monomer, 19 kDa protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 1
|
Biophysical insights from a single chain camelid antibody directed against the Disrupted-in-Schizophrenia 1 protein.
PLoS One 13(1):e0191162 (2018)
Yerabham ASK, Müller-Schiffmann A, Ziehm T, Stadler A, Köber S, Indurkhya X, Marreiros R, Trossbach SV, Bradshaw NJ, Prikulis I, Willbold D, Weiergräber OH, Korth C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Anti-DISC1 single-domain camelid antibody VHH B5 monomer, 14 kDa protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 19
|
Biophysical insights from a single chain camelid antibody directed against the Disrupted-in-Schizophrenia 1 protein.
PLoS One 13(1):e0191162 (2018)
Yerabham ASK, Müller-Schiffmann A, Ziehm T, Stadler A, Köber S, Indurkhya X, Marreiros R, Trossbach SV, Bradshaw NJ, Prikulis I, Willbold D, Weiergräber OH, Korth C
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Anti-DISC1 single-domain camelid antibody VHH B5 monomer, 14 kDa protein
Disrupted- in-schizophrenia 1 (DISC1 12D2) 691-836 monomer, 19 kDa protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 19
|
Biophysical insights from a single chain camelid antibody directed against the Disrupted-in-Schizophrenia 1 protein.
PLoS One 13(1):e0191162 (2018)
Yerabham ASK, Müller-Schiffmann A, Ziehm T, Stadler A, Köber S, Indurkhya X, Marreiros R, Trossbach SV, Bradshaw NJ, Prikulis I, Willbold D, Weiergräber OH, Korth C
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|