|
|
|
|
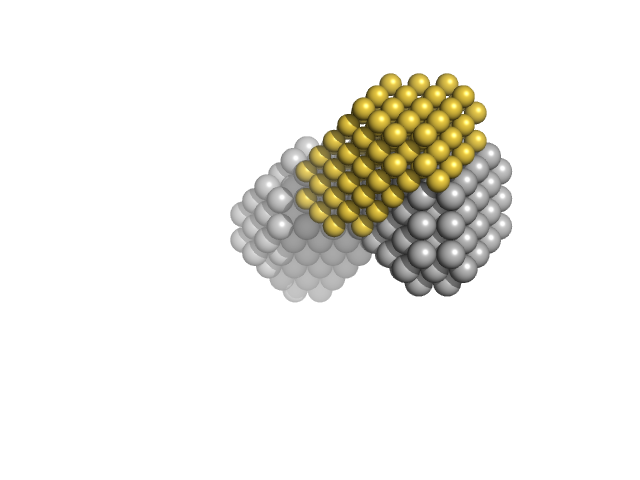
|
| Sample: |
Basic domain of telomeric repeat-binding factor 2 monomer, 5 kDa Homo sapiens protein
Telomere DNA duplex monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 50 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 May 3
|
Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it.
Nucleic Acids Res 45(21):12170-12180 (2017)
Necasová I, Janoušková E, Klumpler T, Hofr C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Telomere DNA duplex monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 50 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 May 3
|
Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it.
Nucleic Acids Res 45(21):12170-12180 (2017)
Necasová I, Janoušková E, Klumpler T, Hofr C
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sortilin 1 A464E alias Neurotensin-receptor 3 A464E monomer, 76 kDa Mus musculus protein
|
| Buffer: |
25 mM MES pH 5.5, 150 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Pomacea maculata perivitellin 1 dodecamer, 278 kDa Pomacea maculata protein
|
| Buffer: |
100 mM Phosphate Buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 Mar 26
|
Convergent evolution of plant and animal embryo defences by hyperstable non-digestible storage proteins.
Sci Rep 7(1):15848 (2017)
Pasquevich MY, Dreon MS, Qiu JW, Mu H, Heras H
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
537 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA polymerase processivity factor component A20 C-ter fragment monomer, 17 kDa Vaccinia virus protein
|
| Buffer: |
25 mM Tris-HCl, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun 8(1):1455 (2017)
Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, Hart DJ, Peyrefitte CN, Burmeister WP, Iseni F
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 16
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
440 |
nm3 |
|
|
|
|
|
|
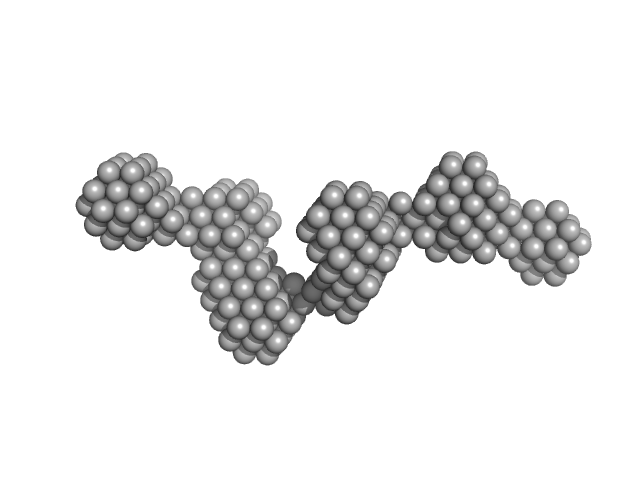
|
| Sample: |
Bacterial cellulose synthesis subunit C monomer, 71 kDa Enterobacter sp. CJF-002 protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Apr 24
|
Crystal structure of the flexible tandem repeat domain of bacterial cellulose synthesis subunit C.
Sci Rep 7(1):13018 (2017)
Nojima S, Fujishima A, Kato K, Ohuchi K, Shimizu N, Yonezawa K, Tajima K, Yao M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
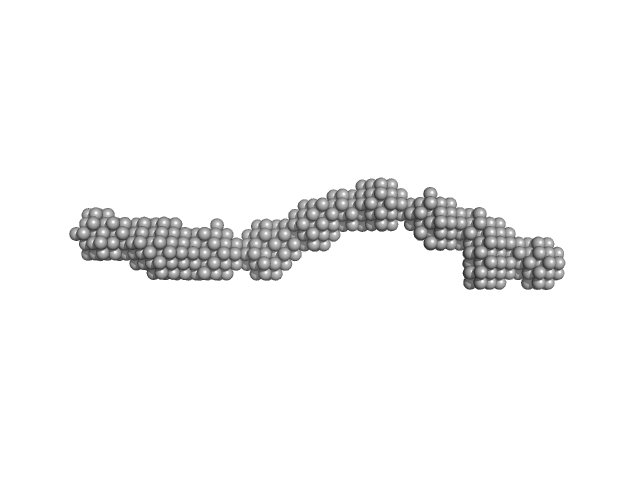
CD22 extracellular domain monomer, 90 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 21
|
Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun 8(1):764 (2017)
Ereño-Orbea J, Sicard T, Cui H, Mazhab-Jafari MT, Benlekbir S, Guarné A, Rubinstein JL, Julien JP
|
| RgGuinier |
8.0 |
nm |
| Dmax |
30.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
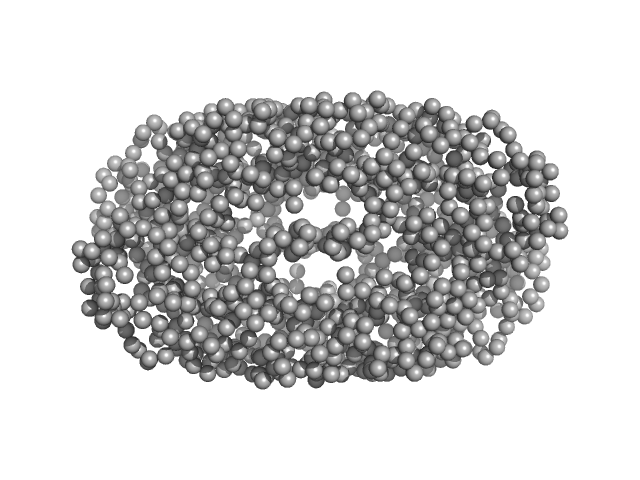
CD22 extracellular domain monomer, 90 kDa Homo sapiens protein
Alpha(2,6)-Sialyllactose, 1 kDa
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 21
|
Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun 8(1):764 (2017)
Ereño-Orbea J, Sicard T, Cui H, Mazhab-Jafari MT, Benlekbir S, Guarné A, Rubinstein JL, Julien JP
|
| RgGuinier |
8.1 |
nm |
| Dmax |
29.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
Geobacillus stearothermophilus DnaB1-300 tetramer, 138 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl and 5 mM β-ME, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 Oct 18
|
Structural analyses of the bacterial primosomal protein DnaB reveal that it is a tetramer and forms a complex with a primosomal re-initiation protein.
J Biol Chem 292(38):15744-15757 (2017)
Li YC, Naveen V, Lin MG, Hsiao CD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
315 |
nm3 |
|
|