|
|
|
|
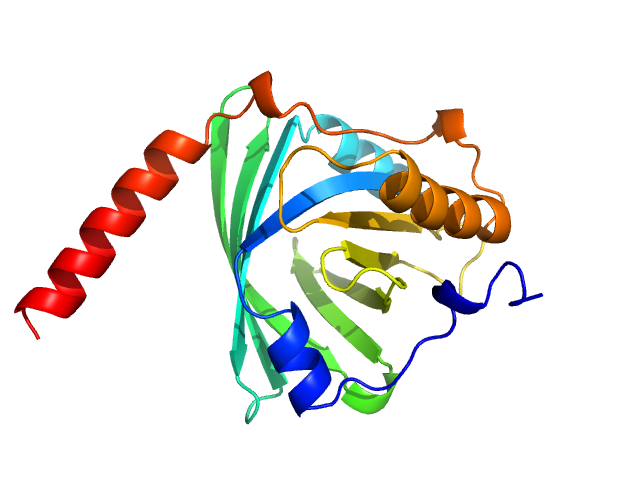
|
| Sample: |
Alpha-1-acid glycoprotein 1 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2021 Jan 12
|
SAXS data based glycosylated models of human alpha-1-acid glycorprotein, a key player in health, disease and drug circulation.
J Biomol Struct Dyn :1-15 (2025)
Kalidas N, Peddada N, Pandey K, Ashish
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Auxin response factor monomer, 46 kDa Marchantia polymorpha protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
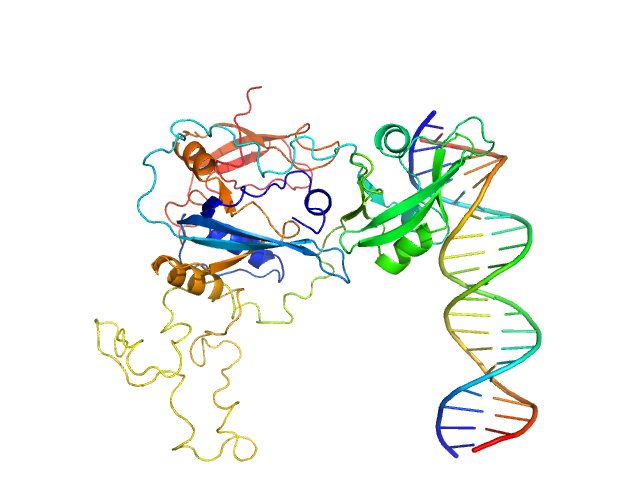
|
| Sample: |
Auxin response factor monomer, 46 kDa Marchantia polymorpha protein
High Affinity ARF binding sequence inverted repeat with 6 nucleotide spacing dimer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
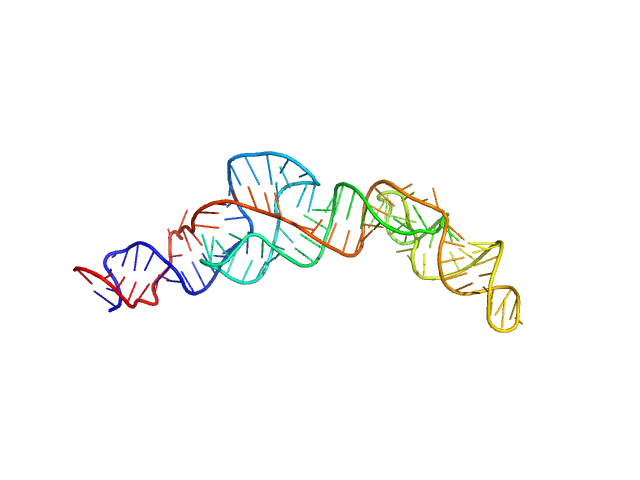
|
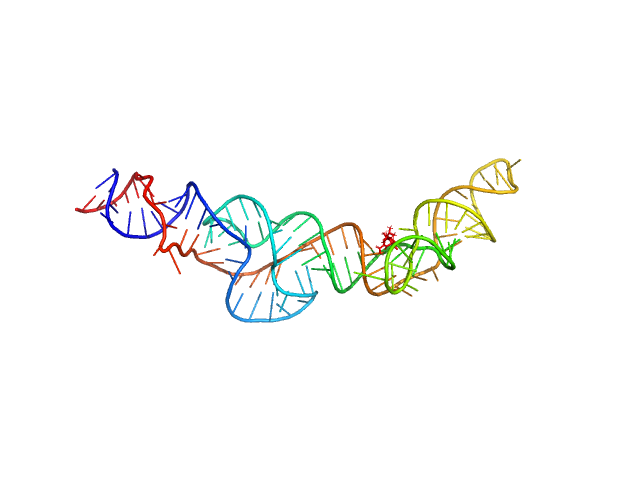
| Sample: |
RNA Device 43 in ligand-free state monomer, 40 kDa synthetic construct RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM KCl, and 1 mM MgCl2, pH: 6.8 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2023 Jul 25
|
Structural investigation of an RNA device that regulates PD-1 expression in mammalian cells.
Nucleic Acids Res 53(5) (2025)
Stagno JR, Deme JC, Dwivedi V, Lee YT, Lee HK, Yu P, Chen SY, Fan L, Degenhardt MFS, Chari R, Young HA, Lea SM, Wang YX
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNA Device 43 in ligand-bound state monomer, 40 kDa synthetic construct RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM KCl, and 1 mM MgCl2, pH: 6.8 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2023 Jul 25
|
Structural investigation of an RNA device that regulates PD-1 expression in mammalian cells.
Nucleic Acids Res 53(5) (2025)
Stagno JR, Deme JC, Dwivedi V, Lee YT, Lee HK, Yu P, Chen SY, Fan L, Degenhardt MFS, Chari R, Young HA, Lea SM, Wang YX
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
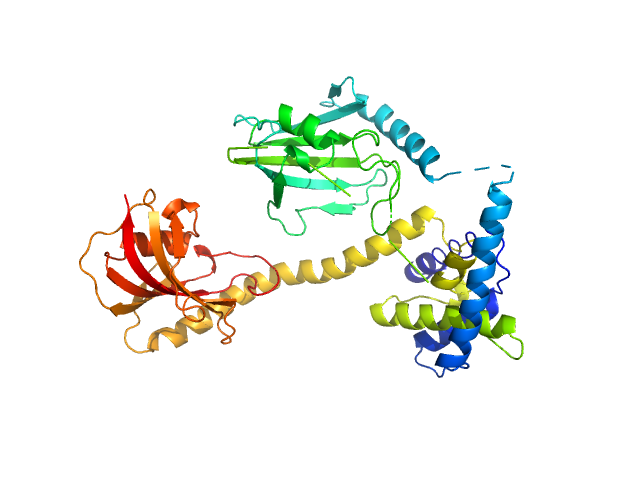
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
(2S)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐4‐methyl‐N‐[(pyridin‐3‐ yl)methyl]pentanamide monomer, 1 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J Med Chem (2025)
Pérez Carrillo VH, Whittaker JJ, Wiedemann C, Harder JM, Lohr T, Jamithireddy AK, Dajka M, Goretzki B, Joseph B, Guskov A, Harmer NJ, Holzgrabe U, Hellmich UA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
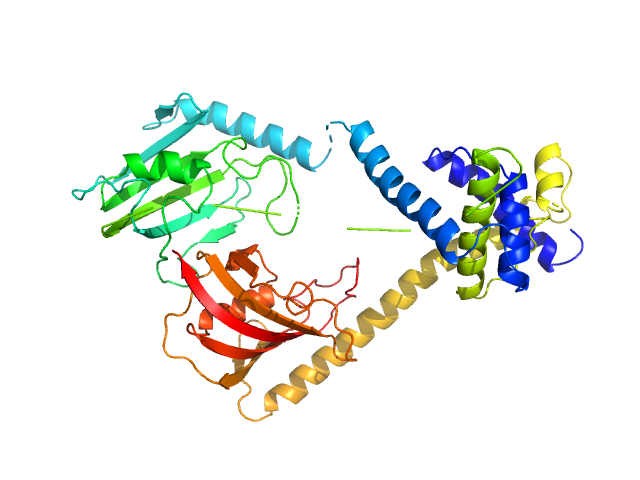
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
(2S)‐3‐(4‐fluorophenyl)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐N‐[(pyridin‐3‐yl)methyl]propanamide monomer, 1 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J Med Chem (2025)
Pérez Carrillo VH, Whittaker JJ, Wiedemann C, Harder JM, Lohr T, Jamithireddy AK, Dajka M, Goretzki B, Joseph B, Guskov A, Harmer NJ, Holzgrabe U, Hellmich UA
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|
|
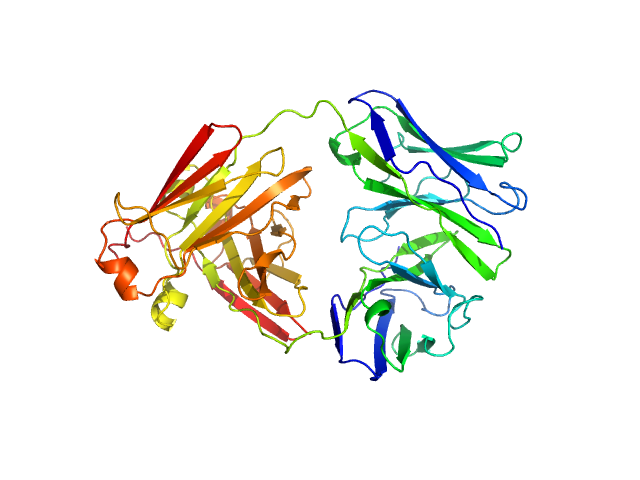
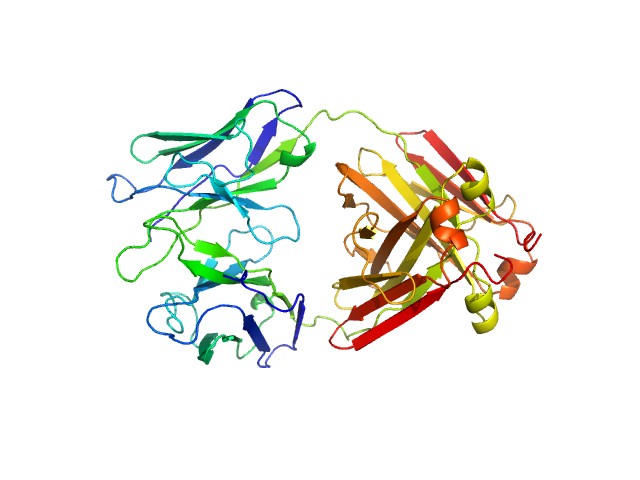
| Sample: |
Immunoglobulin light chain AL55 dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jan 15
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
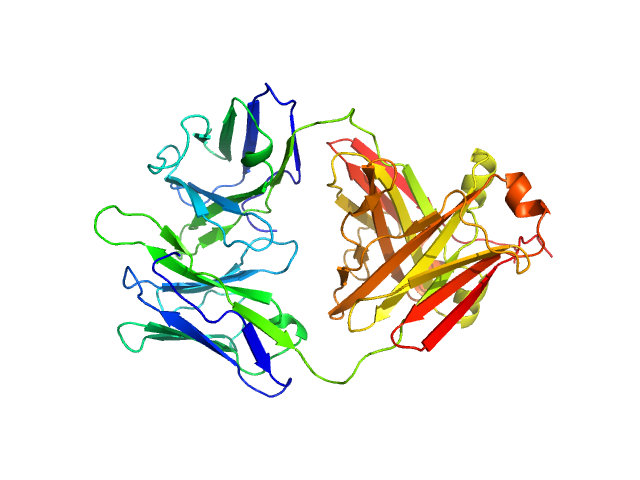
| Sample: |
Immunoglobulin light chain H3 dimer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 12
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Immunoglobulin light chain H7 dimer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 12
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
54 |
nm3 |
|
|