|
|
|
|
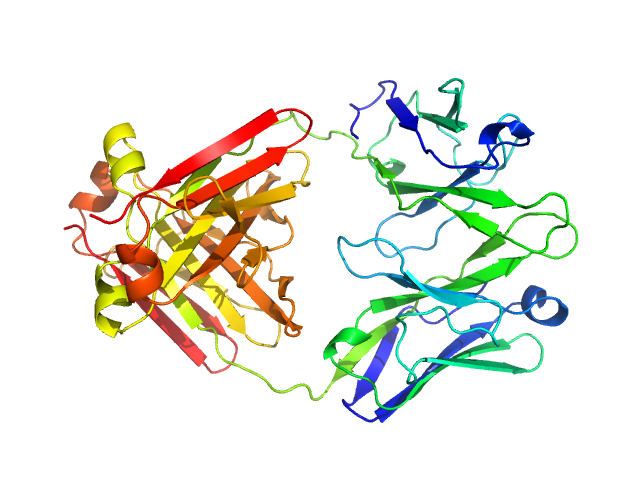
|
| Sample: |
Immunoglobulin light chain H18 dimer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Jul 2
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
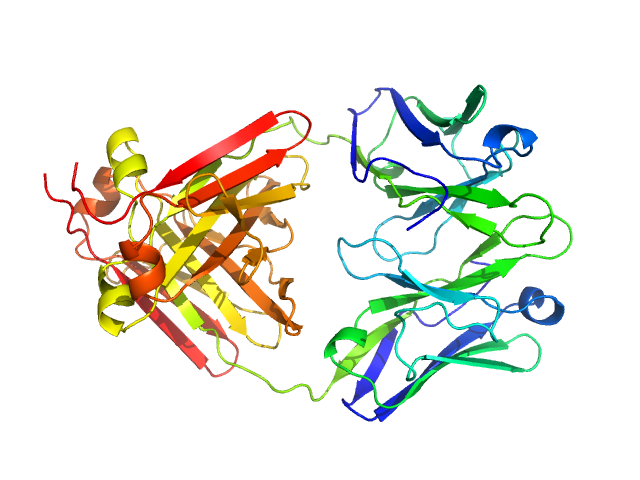
|
| Sample: |
Immunoglobulin light chain M7 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 12
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
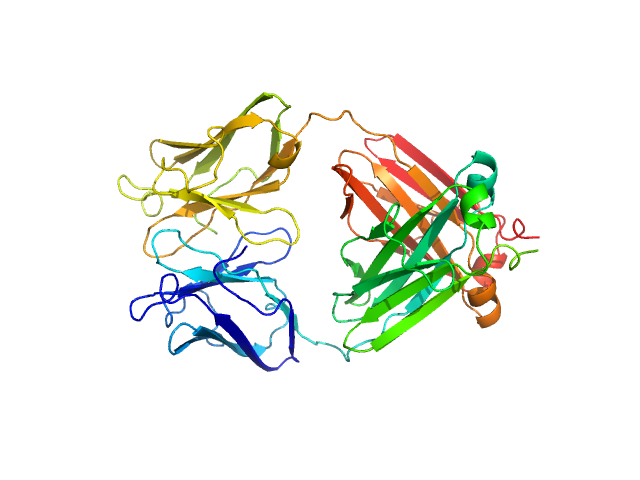
|
| Sample: |
Immunoglobulin light chain M10 dimer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 17
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
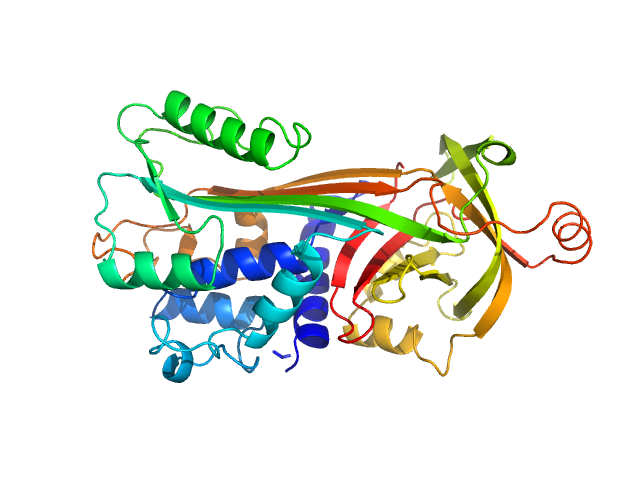
|
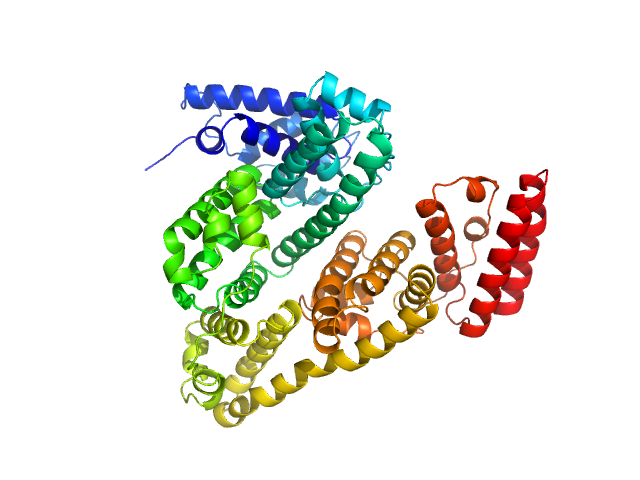
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Albumin (natural variant A214T) monomer, 66 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.7 |
nm |
|
|
|
|
|
|
|
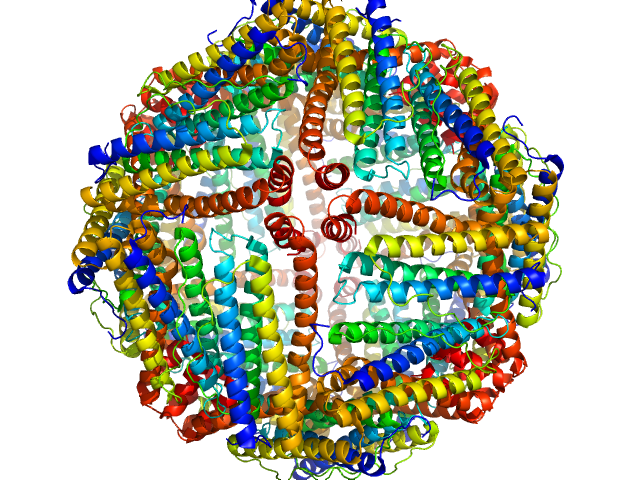
| Sample: |
Ferritin light chain 24-mer, 476 kDa Equus caballus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
23.1 |
nm |
|
|
|
|
|
|
|
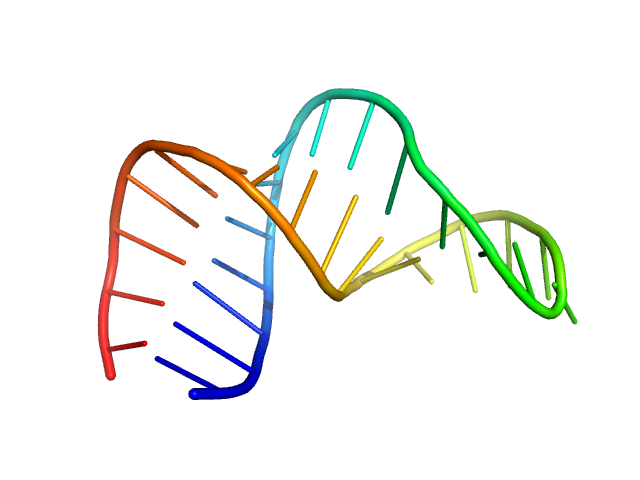
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
|
|
|
|
|
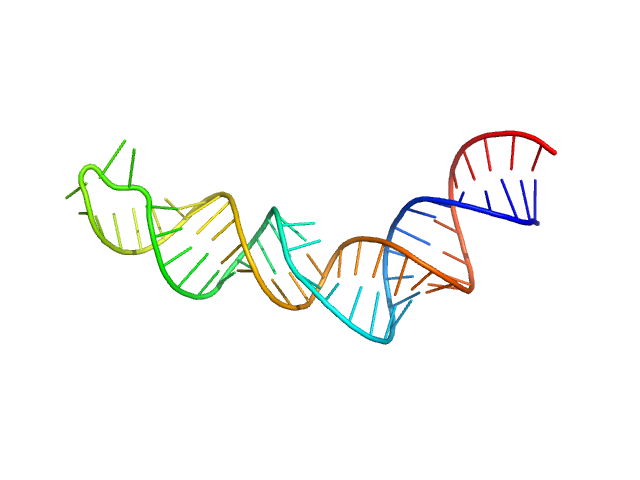
| Sample: |
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
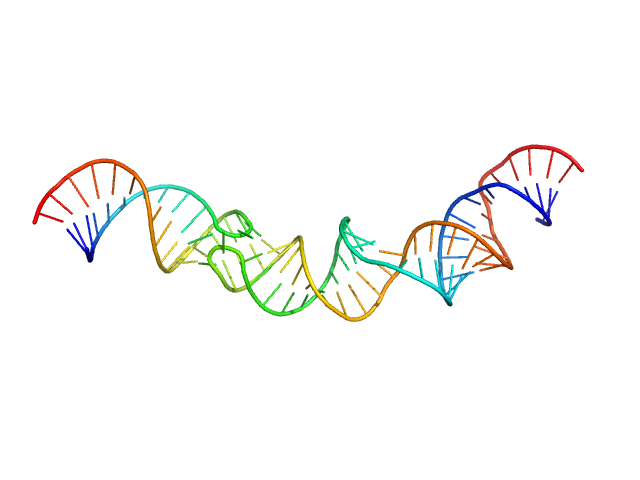
|
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
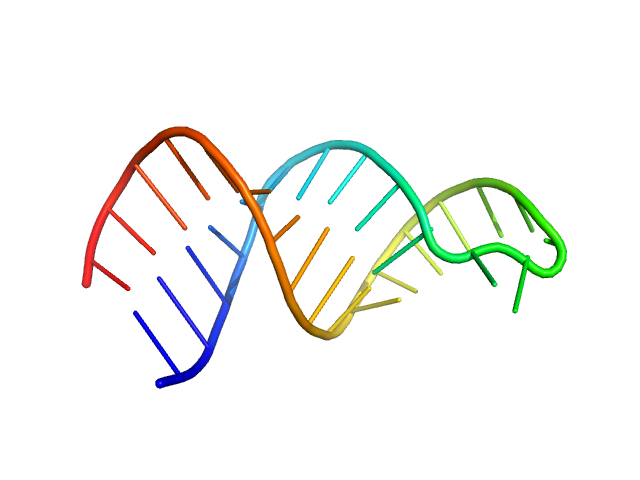
|
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at CG-3, High Flux Isotope Reactor on 2022 Oct 5
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.3 |
nm |
|
|