|
|
|
|
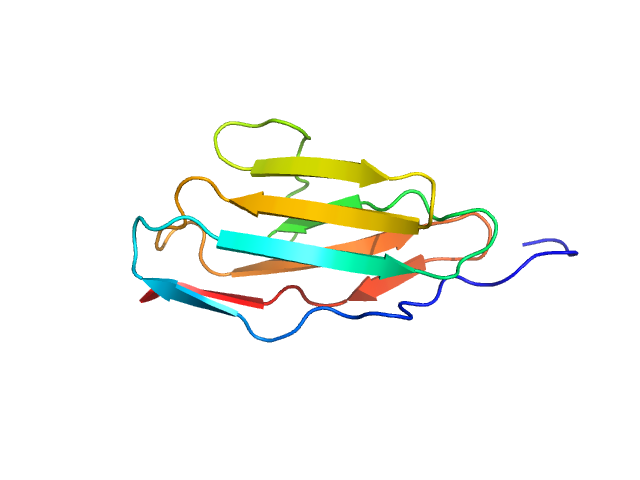
|
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
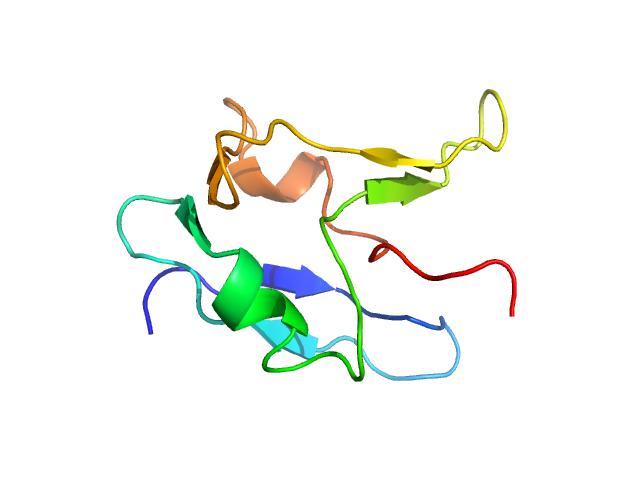
|
| Sample: |
TAR DNA-binding protein 43 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 3% glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BL38B1, SPring-8 on 2024 May 29
|
Semi-synthesis of TDP-43 reveals the effects of phosphorylation in N-terminal domain on self-association
Communications Chemistry 8(1) (2025)
Sasaki D, Tenda M, Sohma Y
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
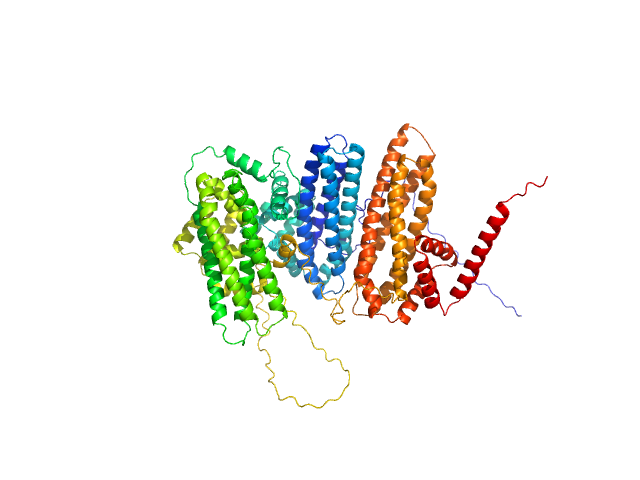
|
| Sample: |
Zinc finger protein BRUTUS-like At1g18910 monomer, 98 kDa Arabidopsis thaliana protein
|
| Buffer: |
10 mM MES, 15 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 8
|
Iron-sensing and redox properties of the hemerythrin-like domains of Arabidopsis BRUTUS and BRUTUS-LIKE2 proteins.
Nat Commun 16(1):3865 (2025)
Pullin J, Rodríguez-Celma J, Franceschetti M, Mundy JEA, Svistunenko DA, Bradley JM, Le Brun NE, Balk J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
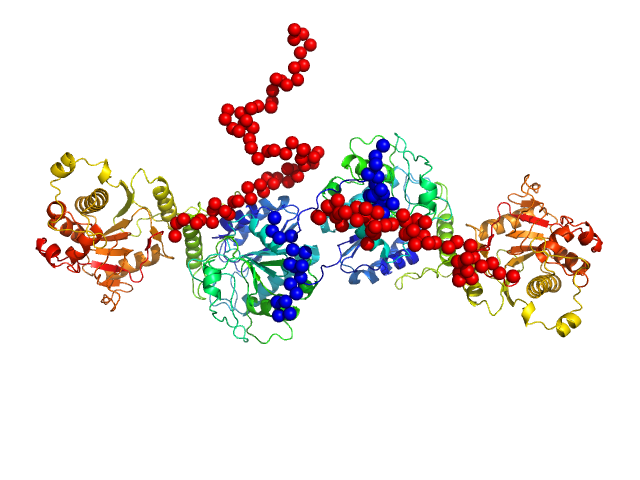
|
| Sample: |
Procollagen galactosyltransferase 1 dimer, 137 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 0.1 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 5
|
Molecular structure and enzymatic mechanism of the human collagen hydroxylysine galactosyltransferase GLT25D1/COLGALT1
Nature Communications 16(1) (2025)
De Marco M, Rai S, Scietti L, Mattoteia D, Liberi S, Moroni E, Pinnola A, Vetrano A, Iacobucci C, Santambrogio C, Colombo G, Forneris F
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
184 |
nm3 |
|
|
|
|
|
|

|
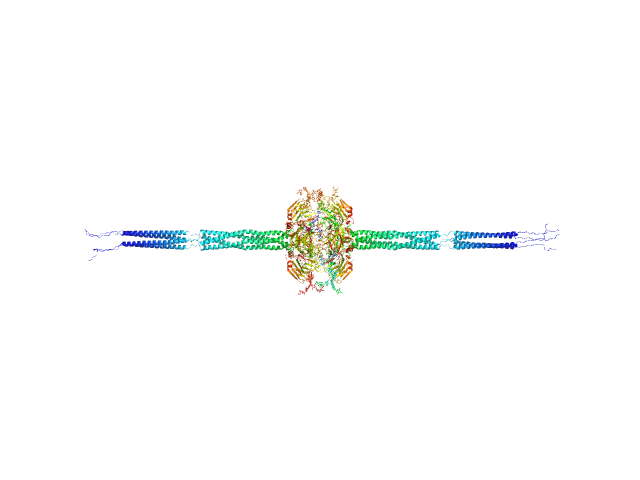
| Sample: |
Pentraxin-related protein PTX3 octamer, 321 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 28
|
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Matrix Biology 136:52-68 (2025)
Shah A, Zhang X, Snee M, Lockhart-Cairns M, Levy C, Jowitt T, Birchenough H, Dean L, Collins R, Dodd R, Roberts A, Enghild J, Mantovani A, Fontana J, Baldock C, Inforzato A, Richter R, Day A
|
| RgGuinier |
8.6 |
nm |
| Dmax |
43.1 |
nm |
| VolumePorod |
658 |
nm3 |
|
|
|
|
|
|
|
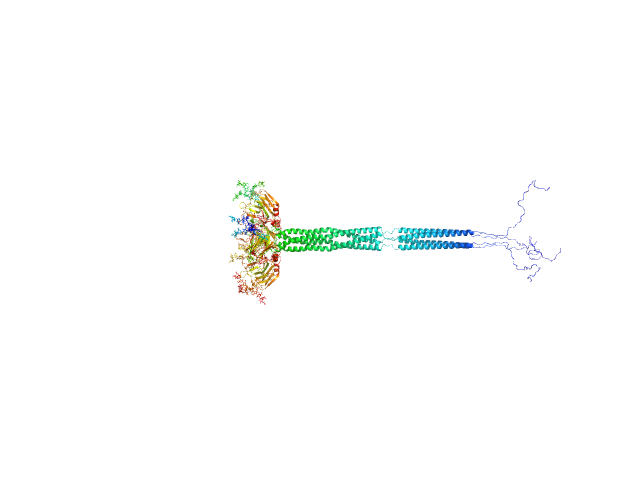
| Sample: |
Pentraxin-related protein PTX3 (N-terminal deletion mutant) octamer, 301 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Matrix Biology 136:52-68 (2025)
Shah A, Zhang X, Snee M, Lockhart-Cairns M, Levy C, Jowitt T, Birchenough H, Dean L, Collins R, Dodd R, Roberts A, Enghild J, Mantovani A, Fontana J, Baldock C, Inforzato A, Richter R, Day A
|
| RgGuinier |
8.4 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
647 |
nm3 |
|
|
|
|
|
|
|
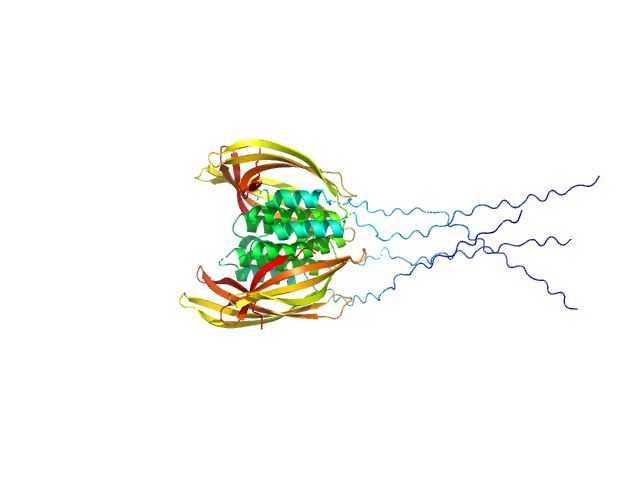
| Sample: |
Pentraxin-related protein PTX3 (C317S, C318S) tetramer, 160 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Matrix Biology 136:52-68 (2025)
Shah A, Zhang X, Snee M, Lockhart-Cairns M, Levy C, Jowitt T, Birchenough H, Dean L, Collins R, Dodd R, Roberts A, Enghild J, Mantovani A, Fontana J, Baldock C, Inforzato A, Richter R, Day A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable conserved Mce associated membrane protein tetramer, 59 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50mM Tris, 500mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 10
|
Insight into the structure and interactions of the M. tuberculosis
Mce-associated membrane proteins Mam1A-1D
(2025)
Hynönen M, Perumal P, Hynönen N, Doutch J, Ma K, Venkatesan R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
90.0 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|

|
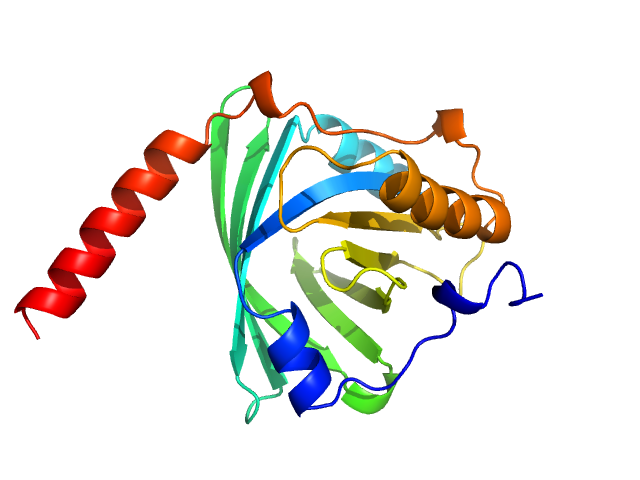
| Sample: |
Probable conserved Mce associated membrane protein tetramer, 59 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, D-C12E9, pH: 8.5 |
| Experiment: |
SANS
data collected at SANS2D, ISIS Neutron and Muon Source on 2019 Dec 10
|
Insight into the structure and interactions of the M. tuberculosis
Mce-associated membrane proteins Mam1A-1D
(2025)
Hynönen M, Perumal P, Hynönen N, Doutch J, Ma K, Venkatesan R
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Alpha-1-acid glycoprotein 1 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2021 Jan 12
|
SAXS data based glycosylated models of human alpha-1-acid glycorprotein, a key player in health, disease and drug circulation.
J Biomol Struct Dyn :1-15 (2025)
Kalidas N, Peddada N, Pandey K, Ashish
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
83 |
nm3 |
|
|