|
|
|
|
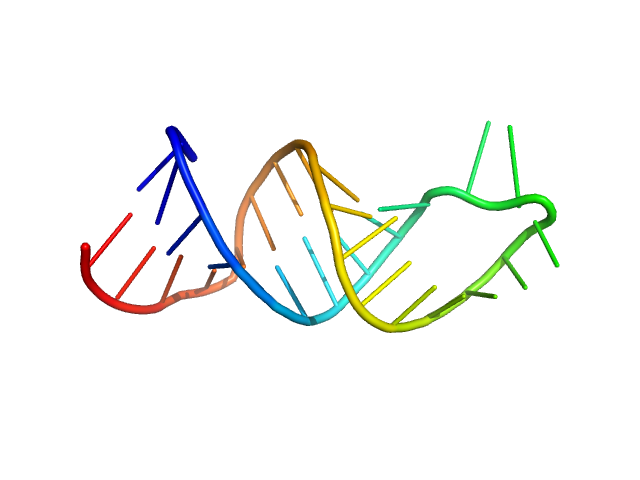
|
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at CG-3, High Flux Isotope Reactor on 2023 Sep 15
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
|
|
|
|
|
|
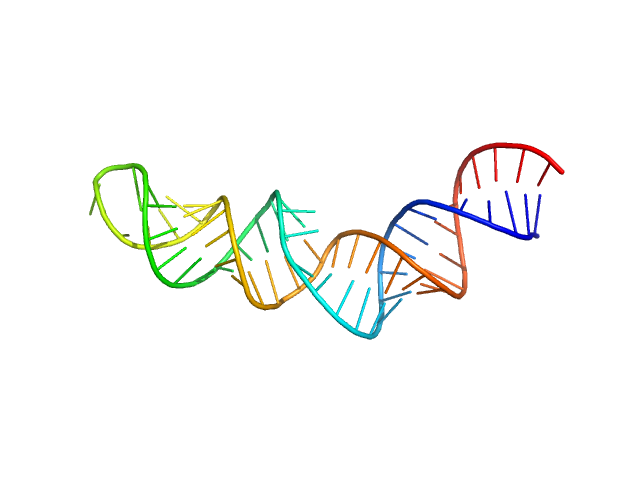
|
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at CG-3, High Flux Isotope Reactor on 2023 Sep 15
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
|
|
|
|
|
|
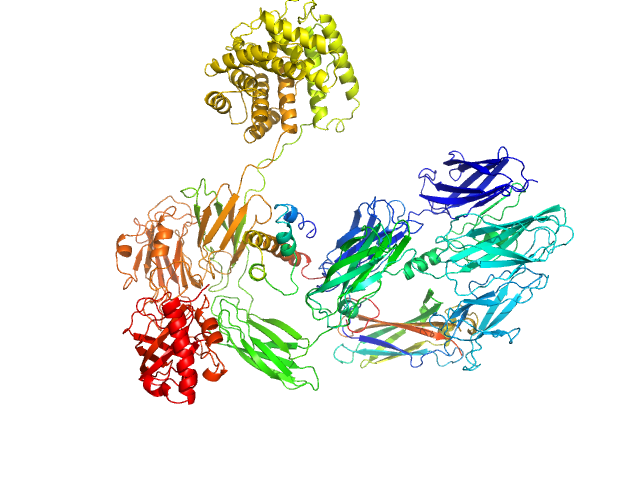
|
| Sample: |
Complement C3 (Δ668-671) monomer, 187 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES pH 6.0, 200 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 7
|
Cryo-EM analysis of complement C3 reveals a reversible major opening of the macroglobulin ring.
Nat Struct Mol Biol (2025)
Gadeberg TAF, Jørgensen MH, Olesen HG, Lorentzen J, Harwood SL, Almeida AV, Fruergaard MU, Jensen RK, Kanis P, Pedersen H, Tranchant E, Petersen SV, Thøgersen IB, Kragelund BB, Lyons JA, Enghild JJ, Andersen GR
|
| RgGuinier |
5.4 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
357 |
nm3 |
|
|
|
|
|
|
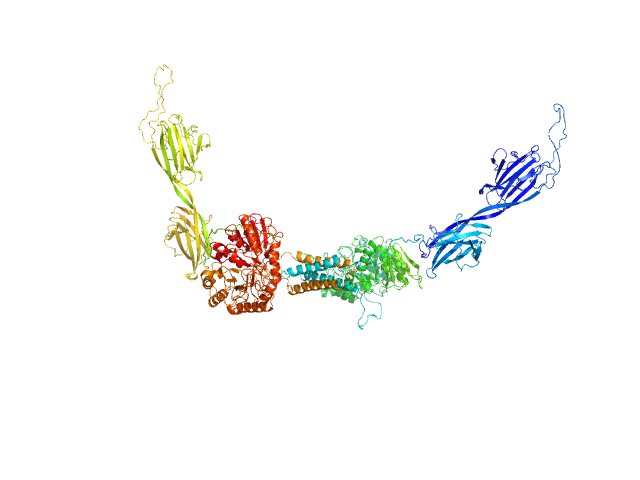
|
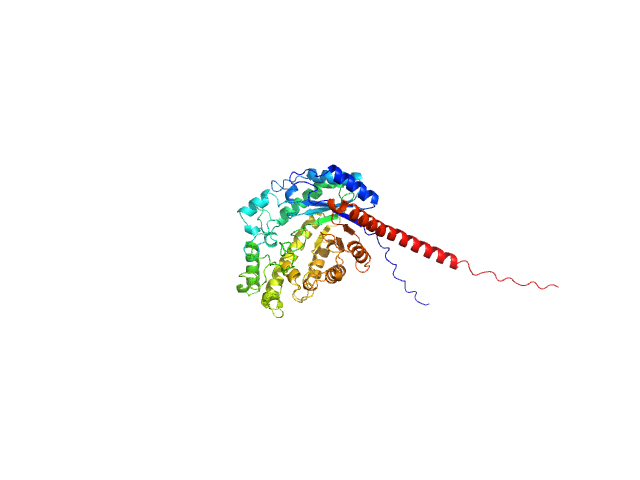
| Sample: |
Alpha-amylase 3, chloroplastic dimer, 187 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
444 |
nm3 |
|
|
|
|
|
|
|
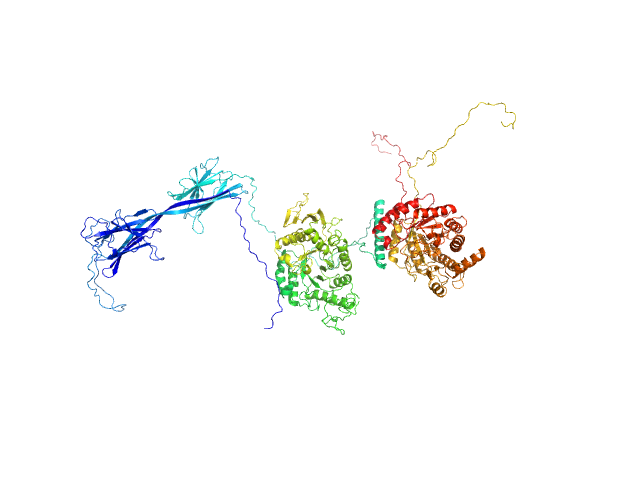
| Sample: |
Inactive beta-amylase 9 monomer, 50 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
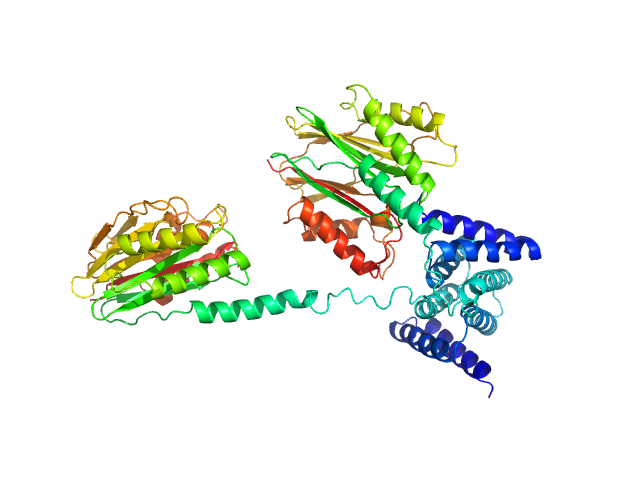
| Sample: |
Inactive beta-amylase 9 monomer, 50 kDa Arabidopsis thaliana protein
Alpha-amylase 3, chloroplastic monomer, 94 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
5.0 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|
|
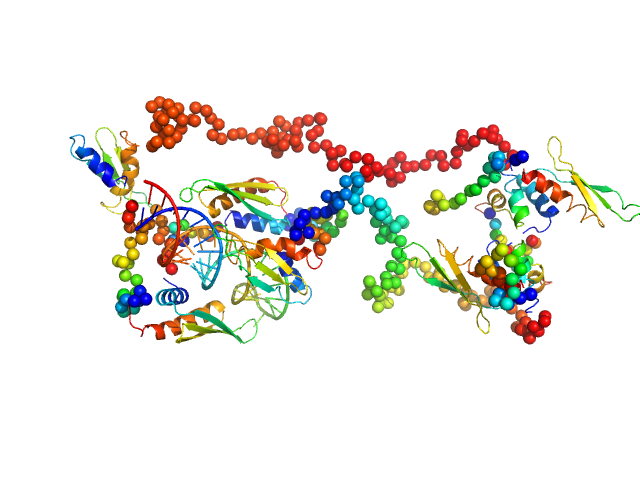
| Sample: |
Phosphoserine phosphatase RsbU dimer, 77 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 24
|
A general mechanism for initiating the bacterial general stress response.
Elife 13 (2025)
Baral R, Ho K, Kumar RP, Hopkins JB, Watkins MB, LaRussa S, Caban-Penix S, Calderone LA, Bradshaw N
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
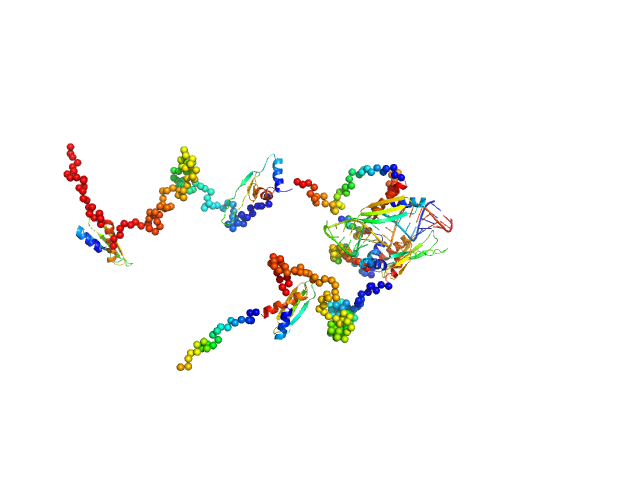
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 (Δ1-177) dimer, 89 kDa Homo sapiens protein
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2020 May 4
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
Double-stranded RNA-binding protein Staufen homolog 1 with truncated RNA-binding domain 2 and truncated Staufen-swapping (ΔSSM) dimer, 81 kDa Homo sapiens protein
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2024 Jan 12
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
5.5 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
|
|
|
|
|
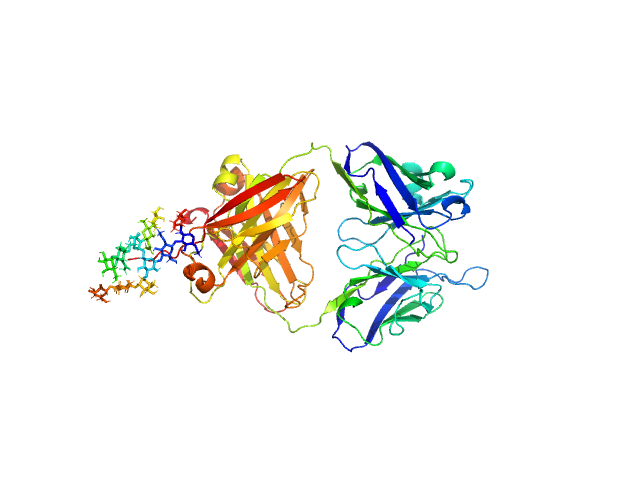
| Sample: |
IgM Mannitou Fab Heavy Chain monomer, 26 kDa Mus musculus protein
IgM Mannitou Fab Light Chain monomer, 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Jun 5
|
Small-angle X-ray scattering of engineered antigen-binding fragments: the case of glycosylated Fab from the Mannitou IgM antibody.
Acta Crystallogr F Struct Biol Commun (2025)
Semwal S, Karamolegkou M, Flament S, Raouraoua N, Verstraete K, Thureau A, Wien F, Bray F, Savvides SN, Bouckaert J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|