|
|
|
|

|
| Sample: |
Phox Homology (PX) - C2 domains of human Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris 200 mM NaCl 5% Glycerol 0.5 mM TCEP, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 20
|
Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2α.
Structure (2018)
Chen KE, Tillu VA, Chandra M, Collins BM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phox Homology (PX) - C2 domains of human Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris 200 mM NaCl 5% Glycerol 0.5 mM TCEP 4 mM InsP6, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 20
|
Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2α.
Structure (2018)
Chen KE, Tillu VA, Chandra M, Collins BM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
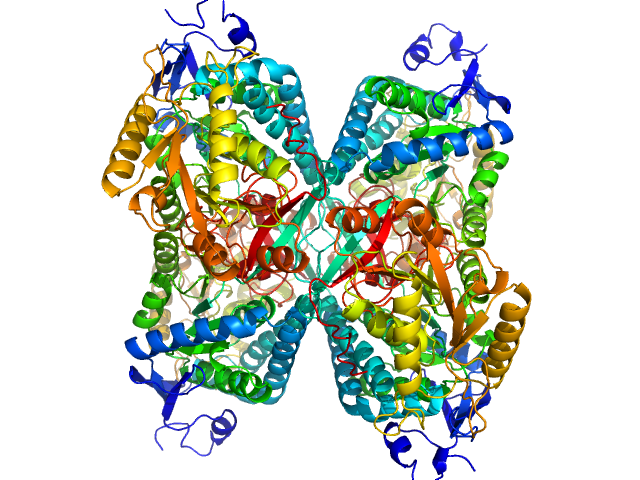
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.5 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
|
|
|
|
|
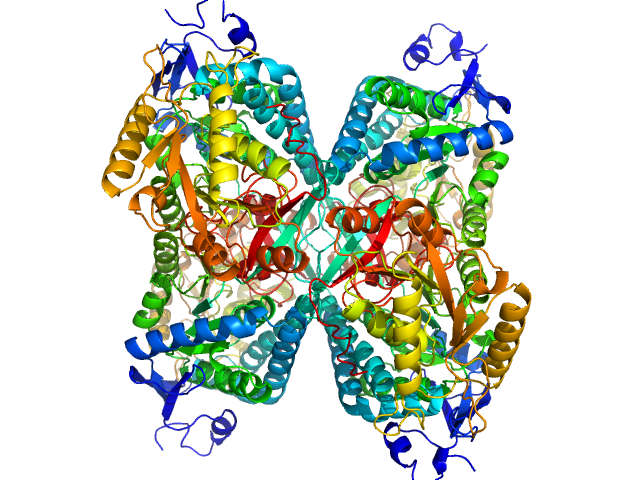
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
229 |
nm3 |
|
|
|
|
|
|
|
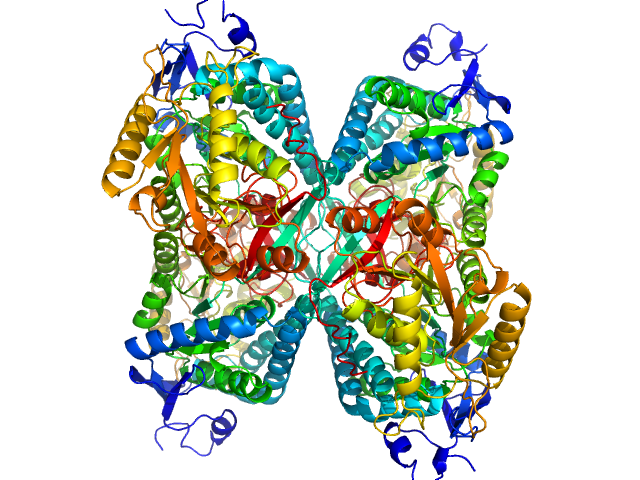
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
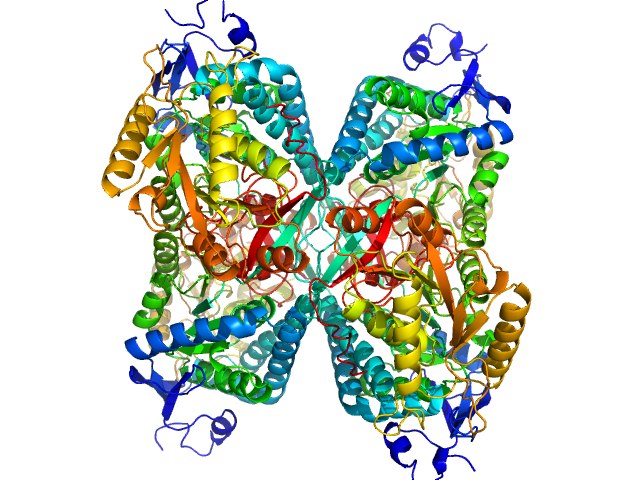
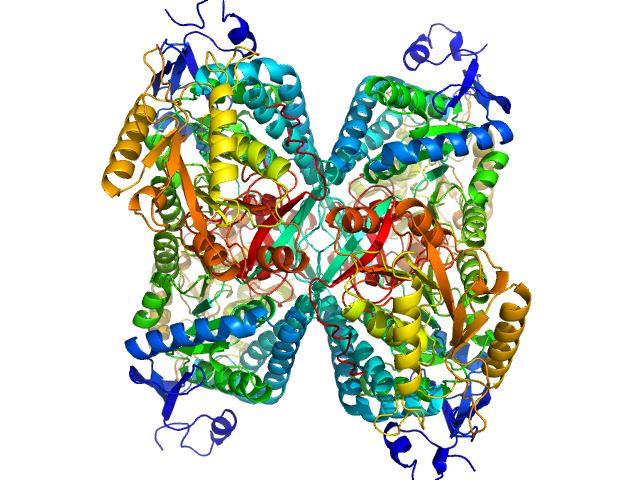
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
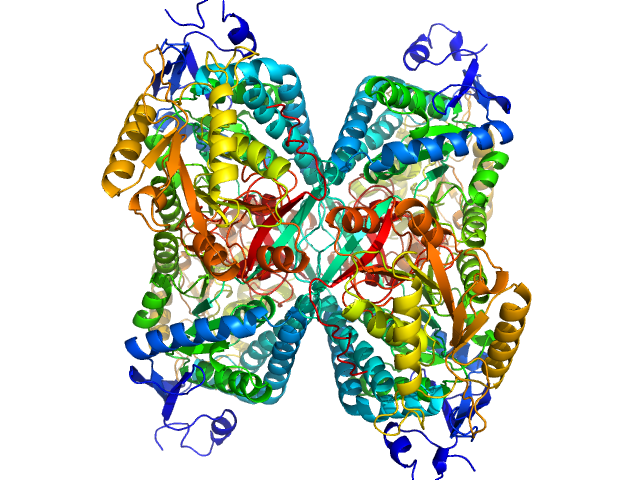
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
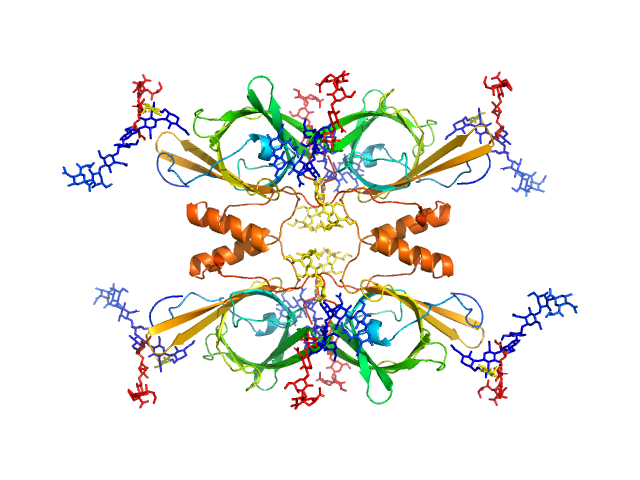
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Identification of a novel tetrameric structure for human apolipoprotein-D.
J Struct Biol 203(3):205-218 (2018)
Kielkopf CS, Low JKK, Mok YF, Bhatia S, Palasovski T, Oakley AJ, Whitten AE, Garner B, Brown SHJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
169 |
nm3 |
|
|
|
|
|
|
|
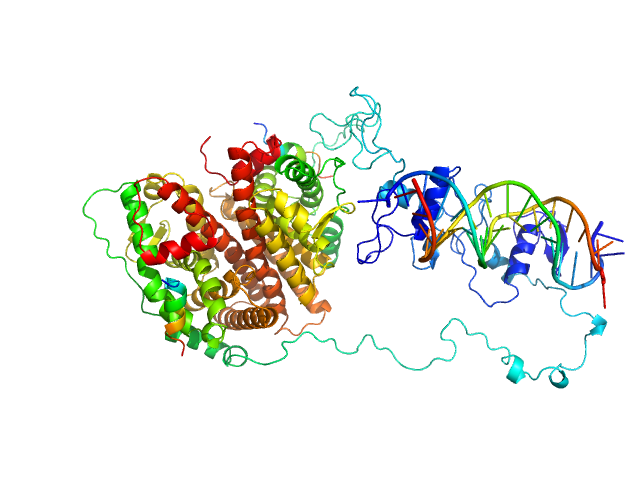
| Sample: |
Estrogen receptor dimer, 85 kDa protein
ERE1 monomer, 6 kDa Homo sapiens DNA
ERE2 monomer, 6 kDa Homo sapiens DNA
Estradiol dimer, 0 kDa
HERa peptide1 monomer, 2 kDa protein
HERa peptide2 monomer, 2 kDa protein
|
| Buffer: |
10 mM CHES (pH9.5), 125 mM NaCl, 5mM KCl, 4 mM MgCl2, 50 mM arginine, 50 mM glutamate, 5 mM TCEP, 5% glycerol, 10 µm Zn acetate, 10 µM estradiol, pH: 9.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Aug 10
|
Multidomain architecture of estrogen receptor reveals interfacial cross-talk between its DNA-binding and ligand-binding domains.
Nat Commun 9(1):3520 (2018)
Huang W, Peng Y, Kiselar J, Zhao X, Albaqami A, Mendez D, Chen Y, Chakravarthy S, Gupta S, Ralston C, Kao HY, Chance MR, Yang S
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
|
|