|
|
|
|
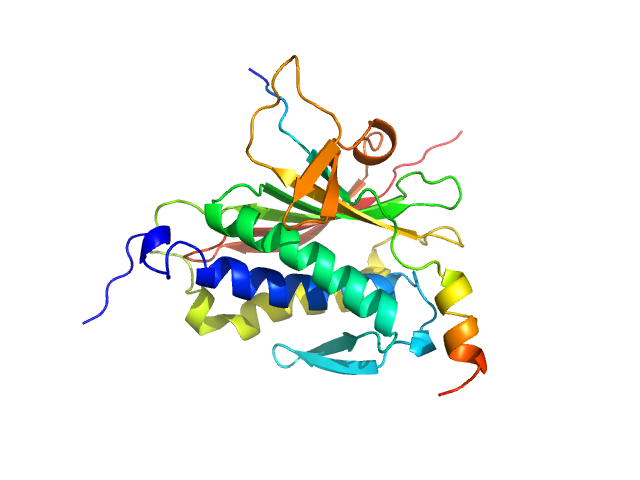
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
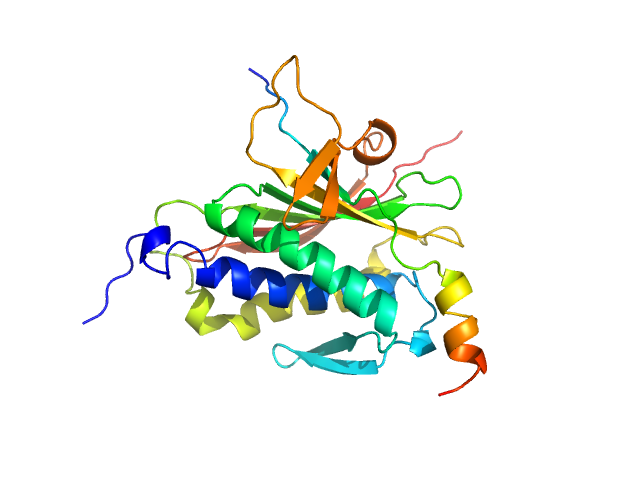
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
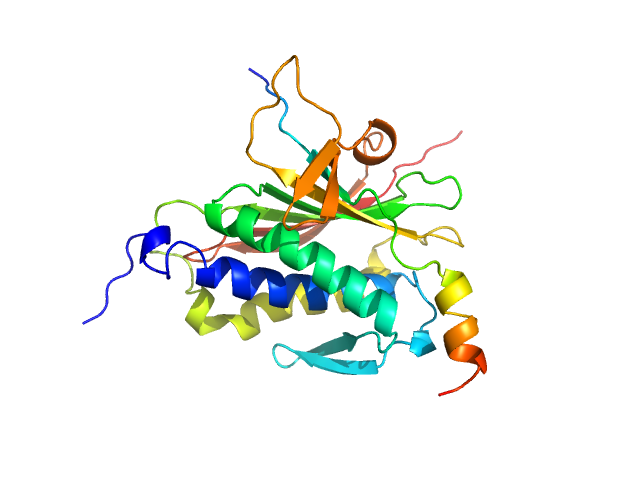
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
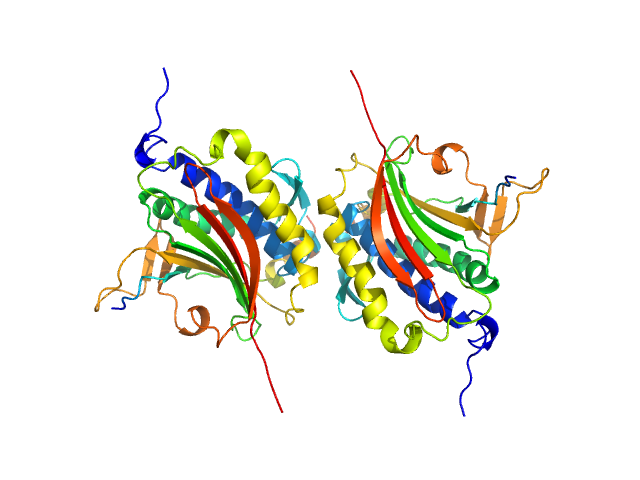
|
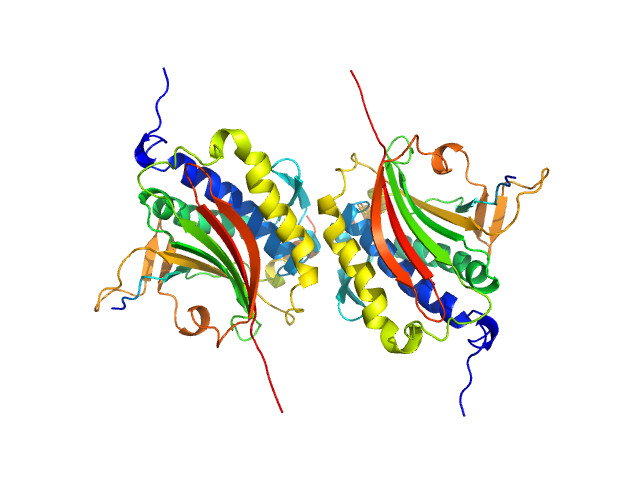
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
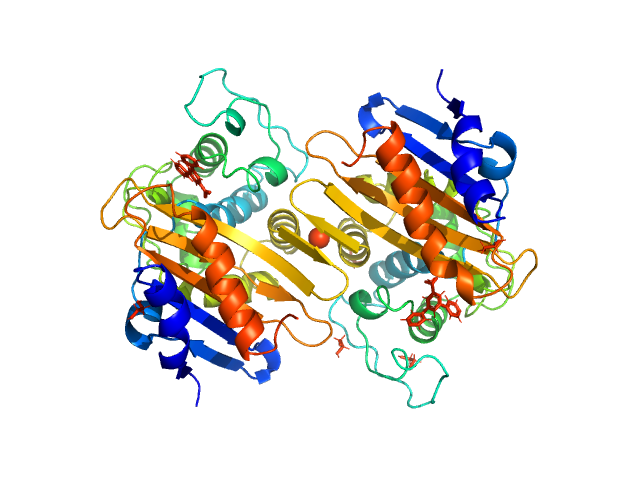
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-lactamase dimer, 56 kDa Klebsiella pneumoniae protein
|
| Buffer: |
50 mM HEPES 50 mM K2SO4, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 23
|
The biological assembly of OXA-48 reveals a dimer interface with high charge complementarity and very high affinity.
FEBS J (2018)
Lund BA, Thomassen AM, Nesheim BHB, Carlsen TJO, Isaksson J, Christopeit T, Leiros HS
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M KCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
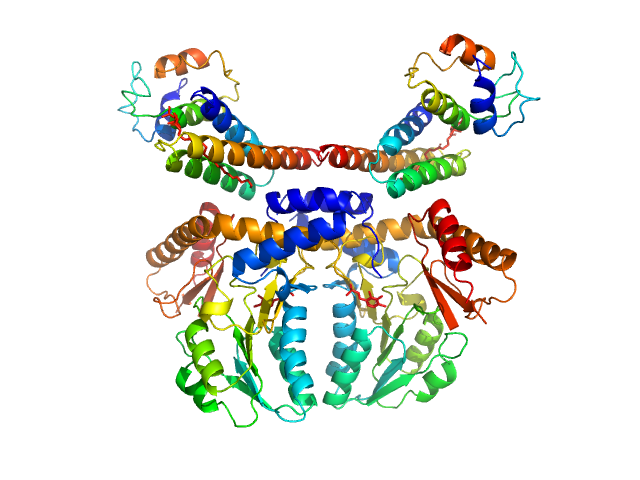
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|
|
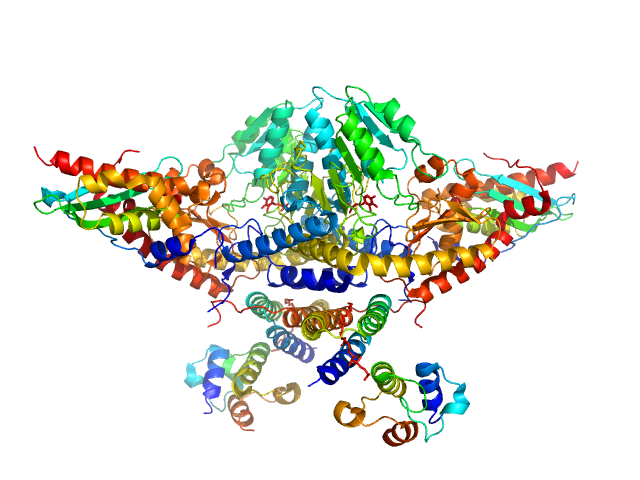
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
Frataxin, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 Apr 18
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
287 |
nm3 |
|
|