|
|
|
|
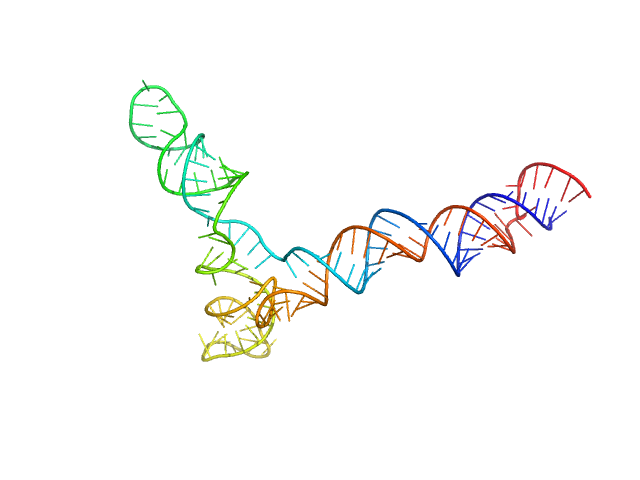
|
| Sample: |
Full stem-loop 5 of SARS-CoV-2 5'genomic end monomer, 48 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.8 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sub-element stem-loop 5a from SARS-CoV-2 5'genomic end monomer, 11 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
|
|
|
|
|
|
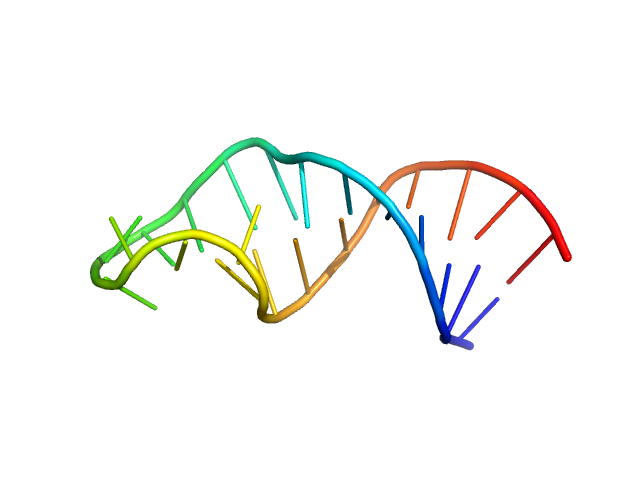
|
| Sample: |
Sub-element stem-loop 5b from SARS-CoV-2 5'genomic end monomer, 8 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Feb 3
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
|
|
|
|
|
|
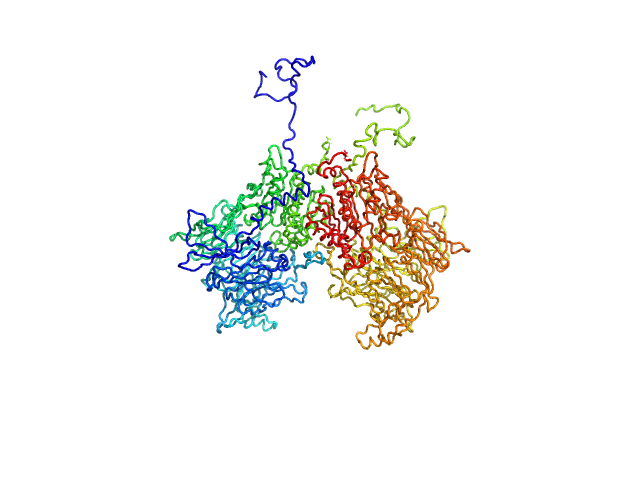
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 8 dimer, 208 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Oct 14
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
271 |
nm3 |
|
|
|
|
|
|
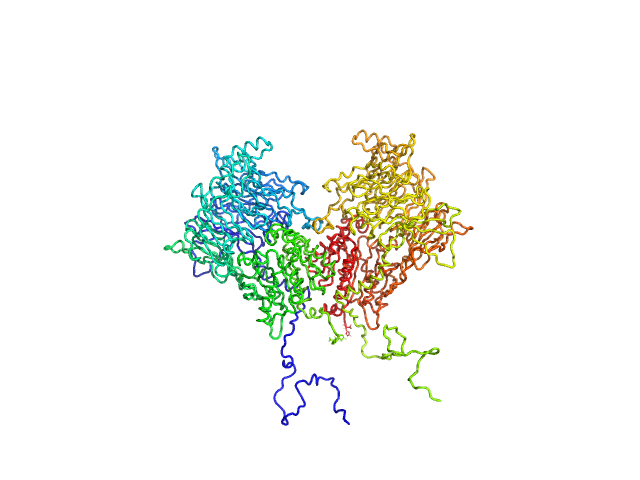
|
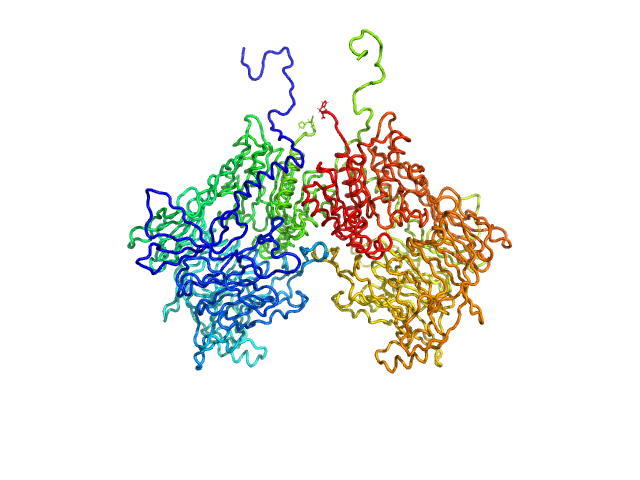
| Sample: |
Isoform 1 of Dipeptidyl peptidase 8 dimer, 208 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Oct 14
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
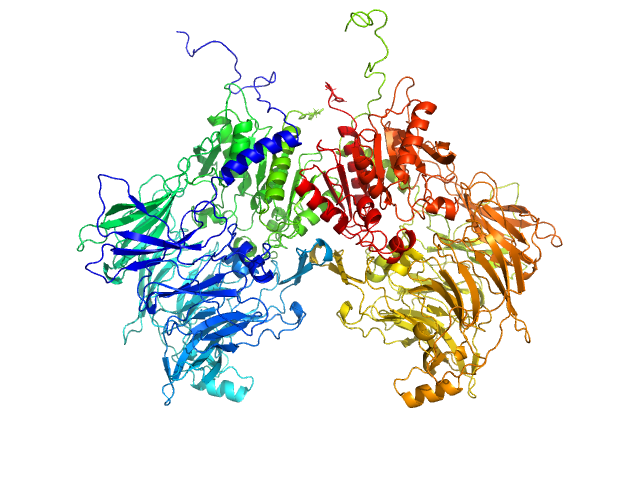
| Sample: |
Isoform 1 of Dipeptidyl peptidase 9 dimer, 198 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
|
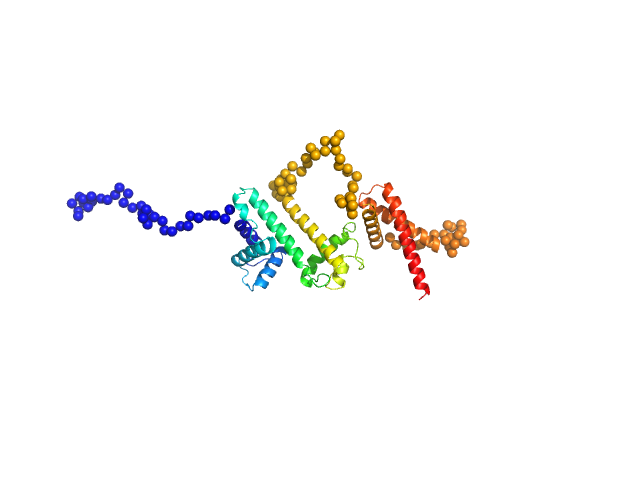
| Sample: |
Isoform 1 of Dipeptidyl peptidase 9 dimer, 198 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
|
|
|
|
|
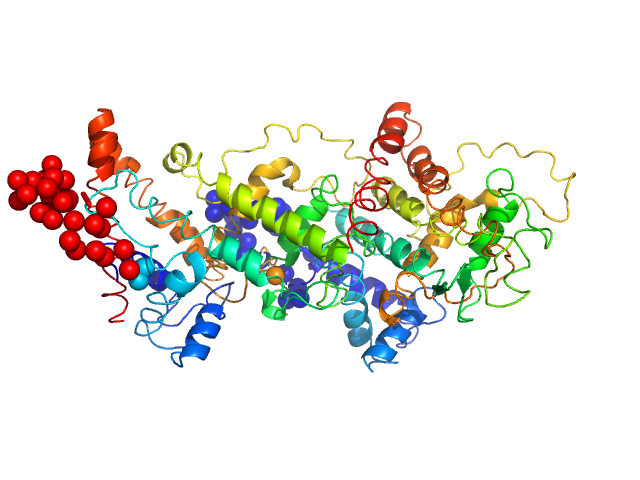
| Sample: |
Harpin Z2 monomer, 37 kDa Pseudomonas syringae strain … protein
|
| Buffer: |
10 mM MES, 100 mM NaF, pH: 6.2 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Centre for Cellular and Molecular Biology on 2023 May 1
|
Solution conformation of HrpZ2 protein from Pseudomonas syringae
Arpita Goswami
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Harpin Z2 monomer, 37 kDa Pseudomonas syringae strain … protein
|
| Buffer: |
10 mM MES, 100 mM NaF, pH: 6.2 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Centre for Cellular and Molecular Biology on 2023 May 1
|
Solution conformation of HrpZ2 protein from Pseudomonas syringae
Arpita Goswami
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
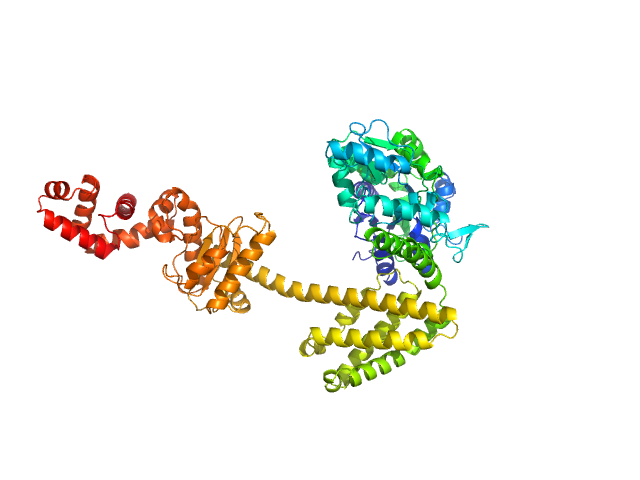
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) monomer, 40 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 monomer, 43 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
113 |
nm3 |
|
|