|
|
|
|
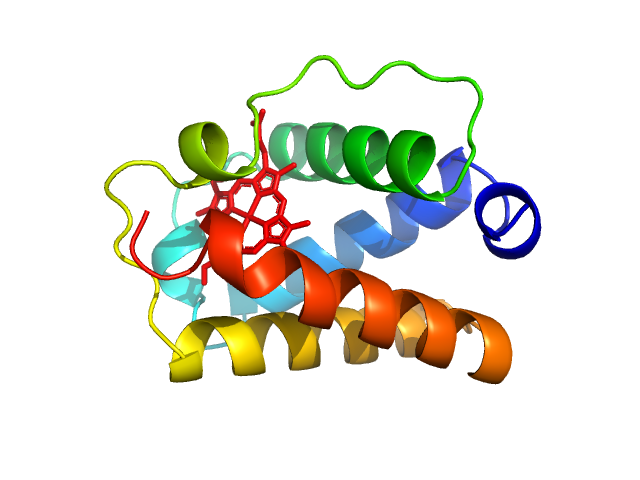
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 May 1
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alpha/beta fold hydrolase monomer, 28 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM NaCl, 10 mM HEPES, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Apr 11
|
Similar but Distinct—Biochemical Characterization of the
Staphylococcus aureus
Serine Hydrolases FphH
and FphI
Proteins: Structure, Function, and Bioinformatics (2024)
Fellner M, Randall G, Bitac I, Warrender A, Sethi A, Jelinek R, Kass I
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
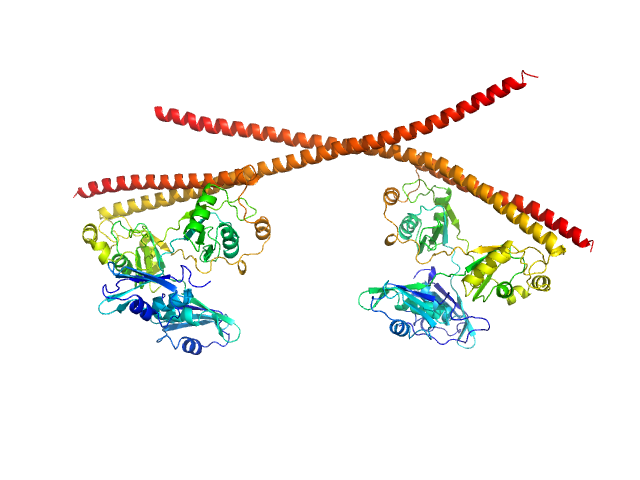
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 18
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
5.5 |
nm |
| Dmax |
20.4 |
nm |
| VolumePorod |
304 |
nm3 |
|
|
|
|
|
|
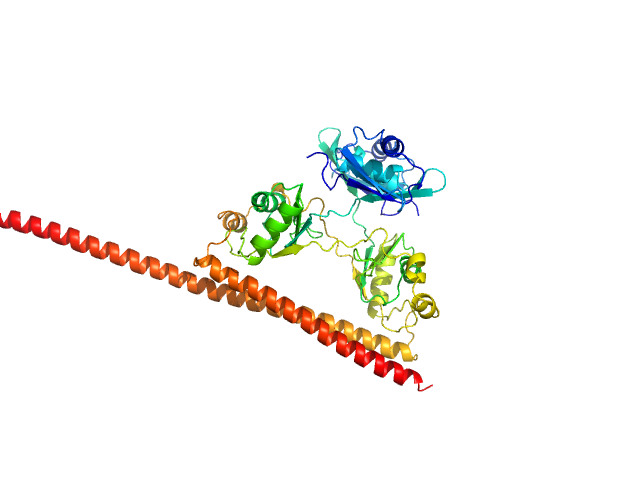
|
| Sample: |
Splicing factor, proline- and glutamine-rich (276-565) monomer, 34 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein (53-312) monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
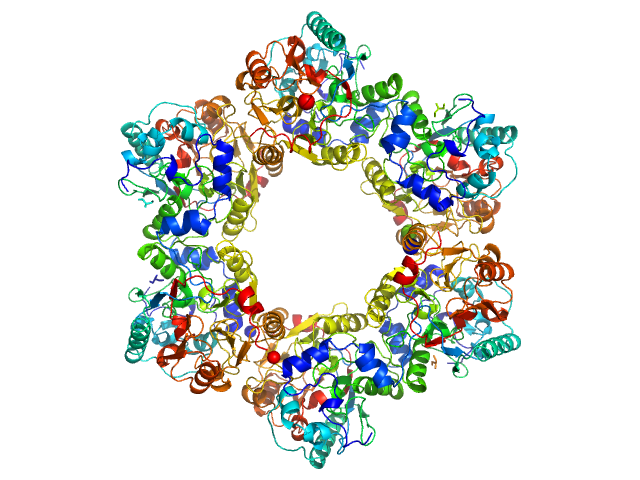
|
| Sample: |
2-nitroimidazole nitrohydrolase (T2I, G14D, K73R) hexamer, 258 kDa Mycobacterium sp. (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% (v/v) glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Aug 6
|
Structural insights into the enzymatic breakdown of azomycin-derived antibiotics by 2-nitroimdazole hydrolase (NnhA).
Commun Biol 7(1):1676 (2024)
Ahmed FH, Liu JW, Royan S, Warden AC, Esquirol L, Pandey G, Newman J, Scott C, Peat TS
|
| RgGuinier |
4.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
363 |
nm3 |
|
|
|
|
|
|
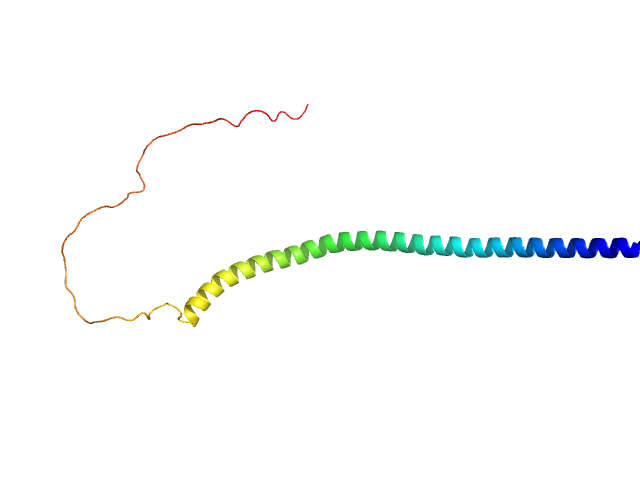
|
| Sample: |
Human alpha-synuclein monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2015 Mar 18
|
Visualizing gaussian-chain like structural models of human α-synuclein in monomeric pre-fibrillar state: Solution SAXS data and modeling analysis.
Int J Biol Macromol 288:138614 (2024)
Dey M, Gupta A, Badmalia MD, Ashish, Sharma D
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
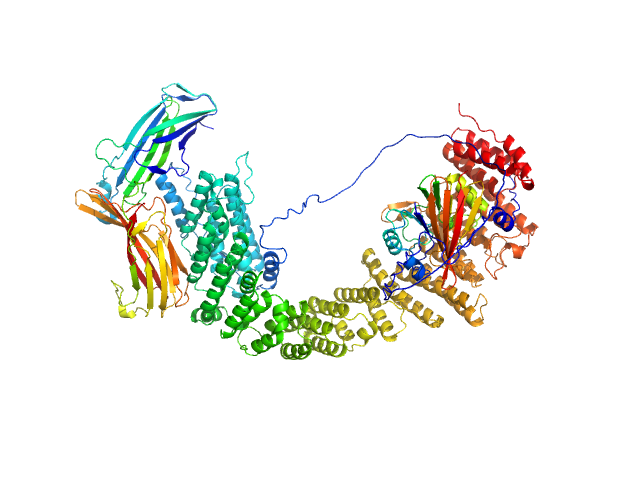
|
| Sample: |
VPS35 endosomal protein-sorting factor-like monomer, 110 kDa Homo sapiens protein
Vacuolar protein sorting-associated protein 29 monomer, 21 kDa Homo sapiens protein
Vacuolar protein sorting-associated protein 26C monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Apr 28
|
Selective cargo and membrane recognition by SNX17 regulates its interaction with Retriever
EMBO Reports (2024)
Martín-González A, Méndez-Guzmán I, Zabala-Zearreta M, Quintanilla A, García-López A, Martínez-Lombardía E, Albesa-Jové D, Acosta J, Lucas M
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
256 |
nm3 |
|
|
|
|
|
|
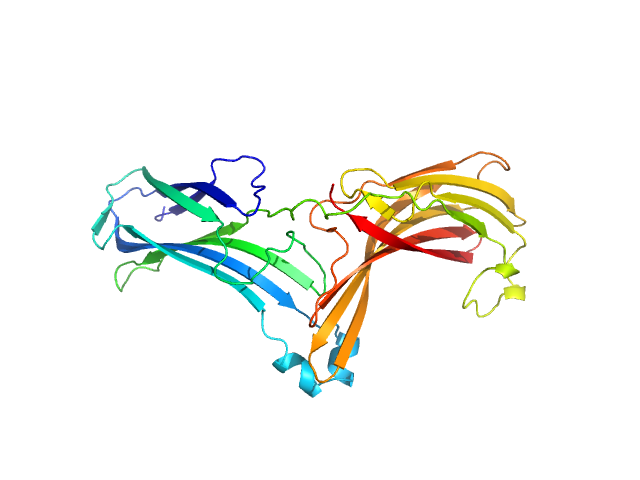
|
| Sample: |
Vacuolar protein sorting-associated protein 26C monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 22
|
Selective cargo and membrane recognition by SNX17 regulates its interaction with Retriever
EMBO Reports (2024)
Martín-González A, Méndez-Guzmán I, Zabala-Zearreta M, Quintanilla A, García-López A, Martínez-Lombardía E, Albesa-Jové D, Acosta J, Lucas M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
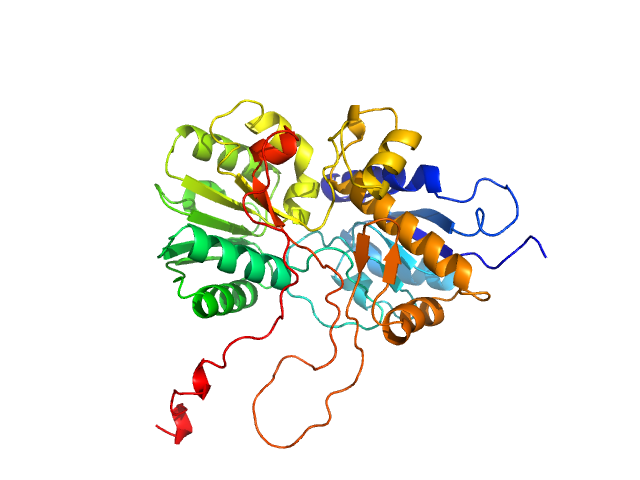
|
| Sample: |
Adenylate cyclase monomer, 43 kDa Trypanosoma brucei protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Biophysical analysis of the membrane-proximal Venus Flytrap domain of ESAG4 receptor-like adenylate cyclase from Trypanosoma brucei.
Mol Biochem Parasitol 260:111653 (2024)
Alves DO, Geens R, da Silva Arruda HR, Jennen L, Corthaut S, Wuyts E, de Andrade GC, Prosdocimi F, Cordeiro Y, Pires JR, Vieira LR, de Oliveira GAP, Sterckx YG, Salmon D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
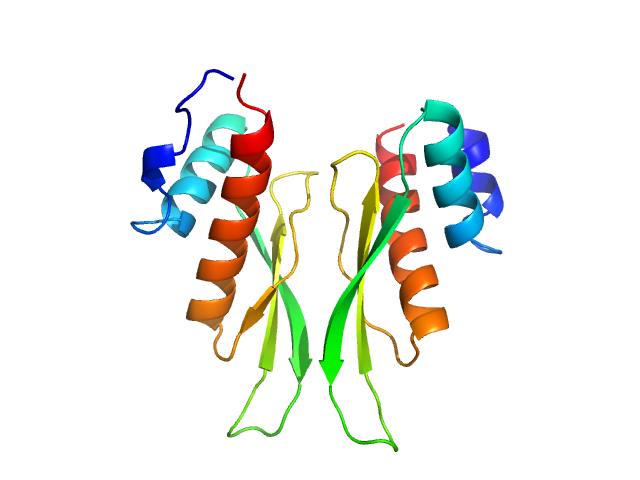
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 716-797, i.e. dsRBD3-short) dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|