|
|
|
|
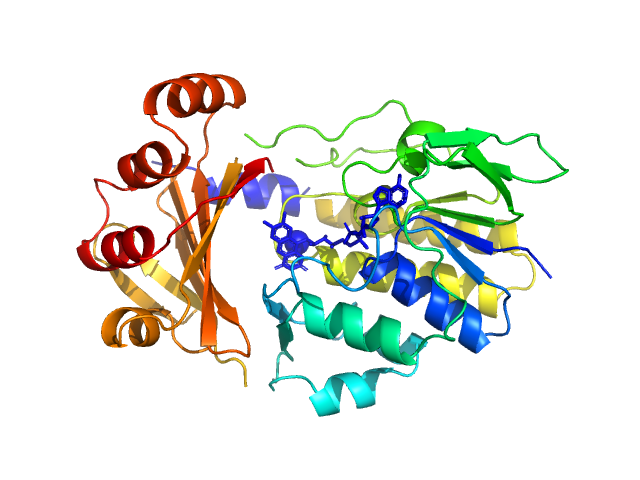
|
| Sample: |
Monooxygenase (M154I, A283T) monomer, 39 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 30 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jan 7
|
Tetracycline-modifying enzyme Sm
TetX from Stenotrophomonas maltophilia
Acta Crystallographica Section F Structural Biology Communications 79(7):180-192 (2023)
Malý M, Kolenko P, Stránský J, Švecová L, Dušková J, Koval' T, Skálová T, Trundová M, Adámková K, Černý J, Božíková P, Dohnálek J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
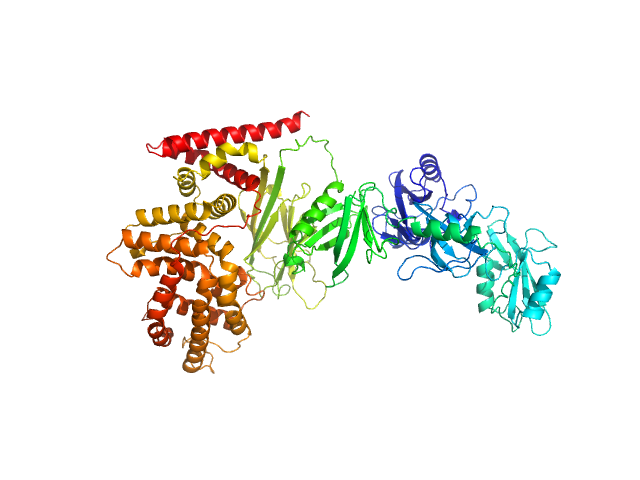
|
| Sample: |
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 4
|
Diverse p120RasGAP interactions with doubly phosphorylated partners EphB4, p190RhoGAP and Dok1
Journal of Biological Chemistry :105098 (2023)
Vish K, Stiegler A, Boggon T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
172 |
nm3 |
|
|
|
|
|
|
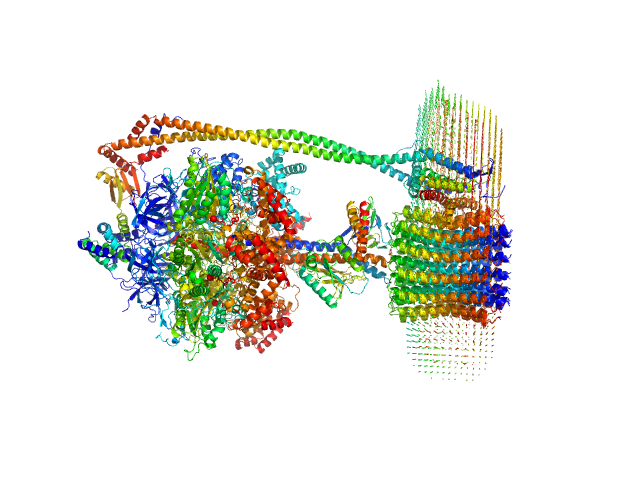
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
300 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
6.6 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
927 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRS8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
150 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
9.6 |
nm |
| Dmax |
41.5 |
nm |
| VolumePorod |
1506 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRT8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
250 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
7.4 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
949 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRU8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
300 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
39.5 |
nm |
| VolumePorod |
1127 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRV8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
350 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
8.9 |
nm |
| Dmax |
44.5 |
nm |
| VolumePorod |
1150 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRW8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
450 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
10.8 |
nm |
| Dmax |
46.5 |
nm |
| VolumePorod |
1703 |
nm3 |
|
|
|
|
|
|
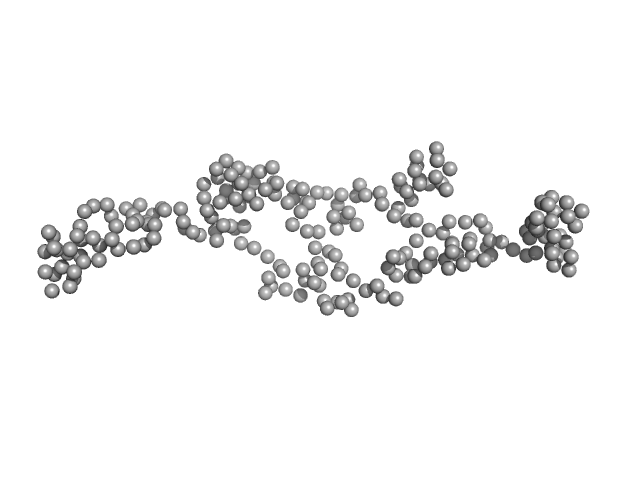
|
| Sample: |
AT5g04600/T32M21_200 monomer, 25 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HNa2PO4, 300 mM NaCl, 5% glycerol (v/v), 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 22
|
Structural and functional analysis of a plant nucleolar RNA chaperone-like protein.
Sci Rep 13(1):9656 (2023)
Fernandes R, Ostendorp A, Ostendorp S, Mehrmann J, Falke S, Graewert MA, Weingartner M, Kehr J, Hoth S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
3-phosphoinositide-dependent protein kinase 1 monomer, 65 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl pH 7.4, 250 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 26
|
Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein
Science Signaling 16(789) (2023)
Sacerdoti M, Gross L, Riley A, Zehnder K, Ghode A, Klinke S, Anand G, Paris K, Winkel A, Herbrand A, Godage H, Cozier G, Süß E, Schulze J, Pastor-Flores D, Bollini M, Cappellari M, Svergun D, Gräwert M, Aramendia P, Leroux A, Potter B, Camacho C, Biondi R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
107 |
nm3 |
|
|