|
|
|
|

|
| Sample: |
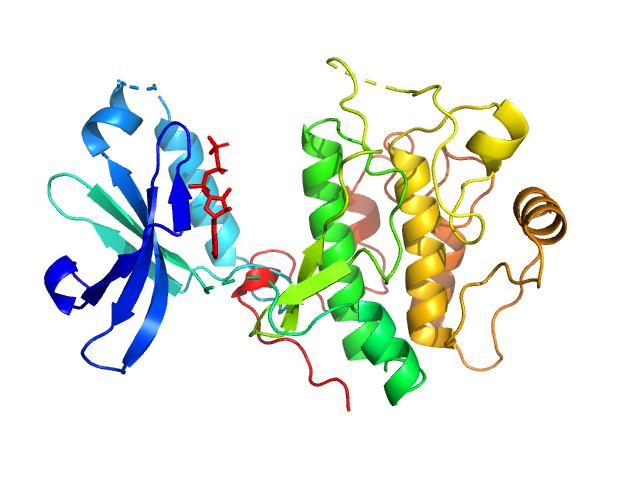
3-phosphoinositide-dependent protein kinase 1 monomer, 65 kDa Homo sapiens protein
2-O-benzoyl-Ins(1,3,4,5,6)P5 monomer, 1 kDa synthetic construct
|
| Buffer: |
20 mM Tris-HCl pH 7.4, 250 mM NaCl, 1 mM DTT, 1 μM HYG8, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 26
|
Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein
Science Signaling 16(789) (2023)
Sacerdoti M, Gross L, Riley A, Zehnder K, Ghode A, Klinke S, Anand G, Paris K, Winkel A, Herbrand A, Godage H, Cozier G, Süß E, Schulze J, Pastor-Flores D, Bollini M, Cappellari M, Svergun D, Gräwert M, Aramendia P, Leroux A, Potter B, Camacho C, Biondi R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
3-phosphoinositide-dependent protein kinase 1 (Y188G Q292A) monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl pH 7.4, 250 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 26
|
Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein
Science Signaling 16(789) (2023)
Sacerdoti M, Gross L, Riley A, Zehnder K, Ghode A, Klinke S, Anand G, Paris K, Winkel A, Herbrand A, Godage H, Cozier G, Süß E, Schulze J, Pastor-Flores D, Bollini M, Cappellari M, Svergun D, Gräwert M, Aramendia P, Leroux A, Potter B, Camacho C, Biondi R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
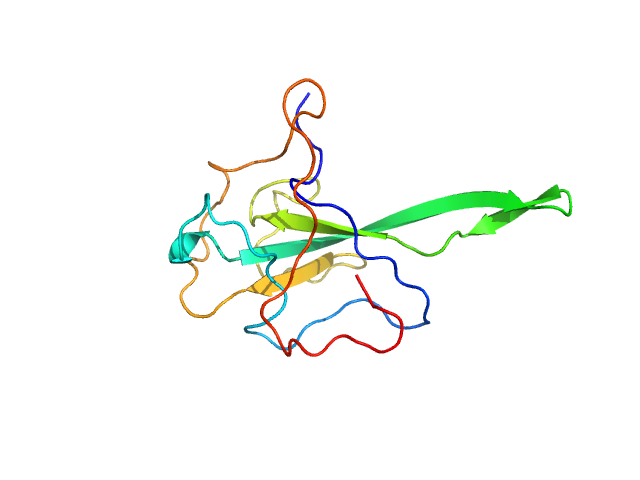
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 16
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
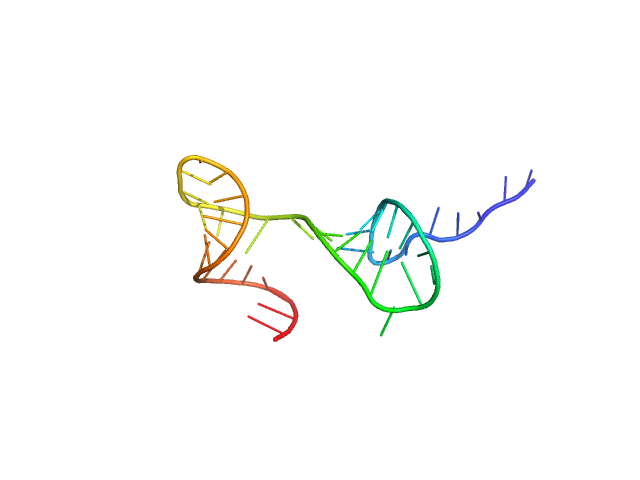
| Sample: |
Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
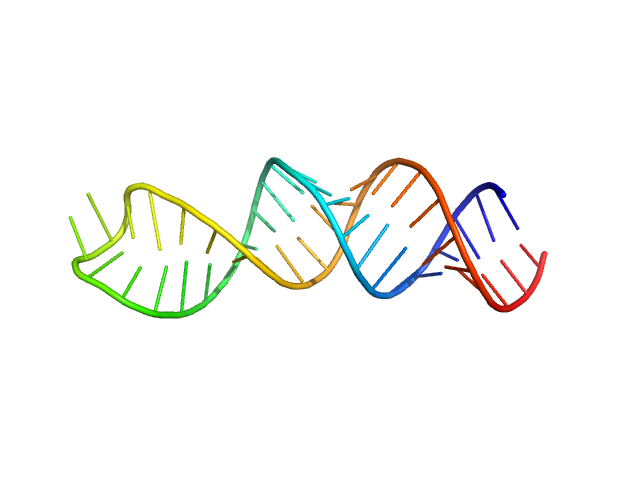
| Sample: |
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 7 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
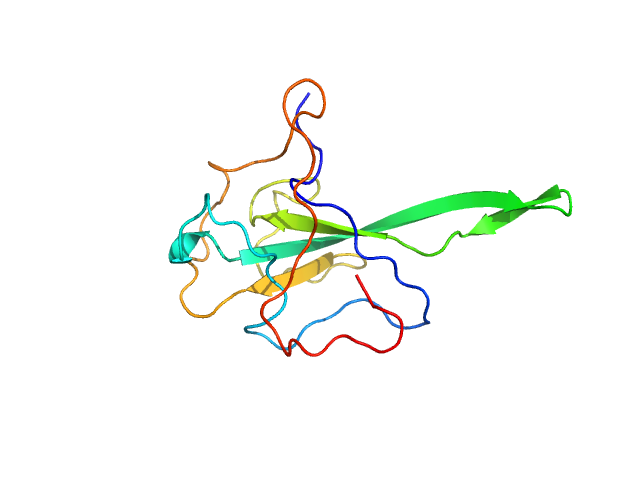
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
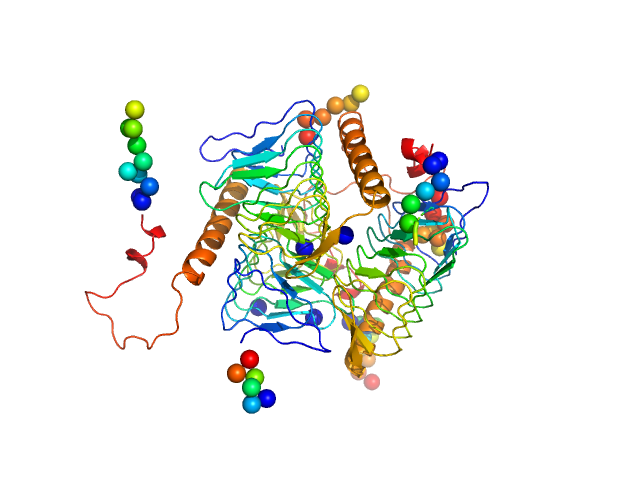
| Sample: |
Bacterial transferase hexapeptide repeat protein trimer, 66 kDa Acinetobacter baumannii (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jan 29
|
Mechanistic and structural insights into the bifunctional enzyme PaaY from Acinetobacter baumannii.
Structure (2023)
Jiao M, He W, Ouyang Z, Qin Q, Guo Y, Zhang J, Bai Y, Guo X, Yu Q, She J, Hwang PM, Zheng F, Wen Y
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
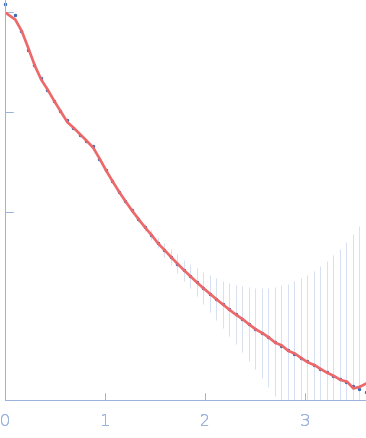
|
| Sample: |
Recombination protein 107 tetramer, 139 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
8.3 |
nm |
| Dmax |
32.2 |
nm |
|
|