|
|
|
|
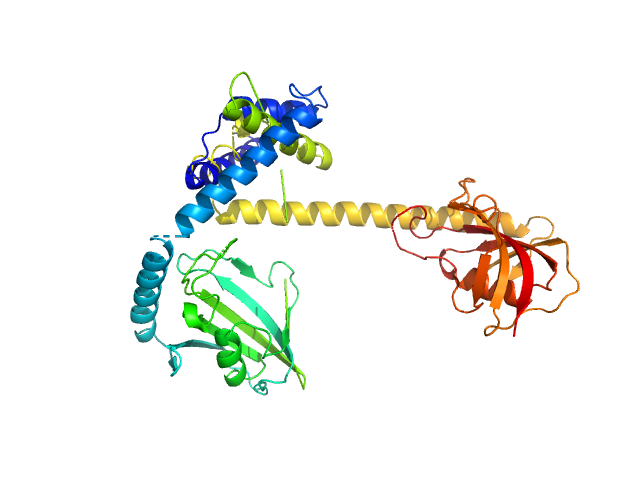
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
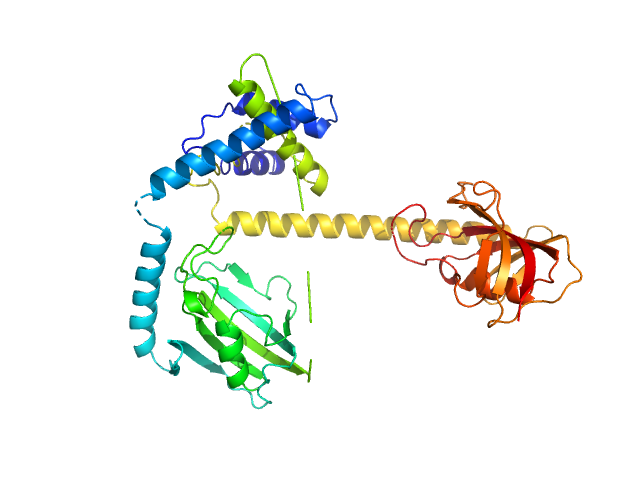
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|
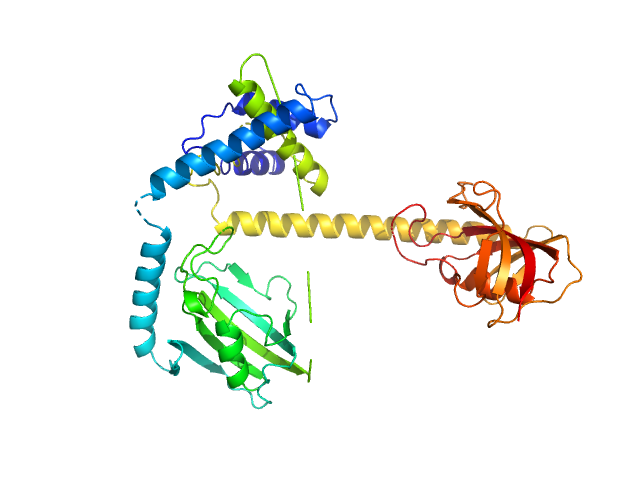
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
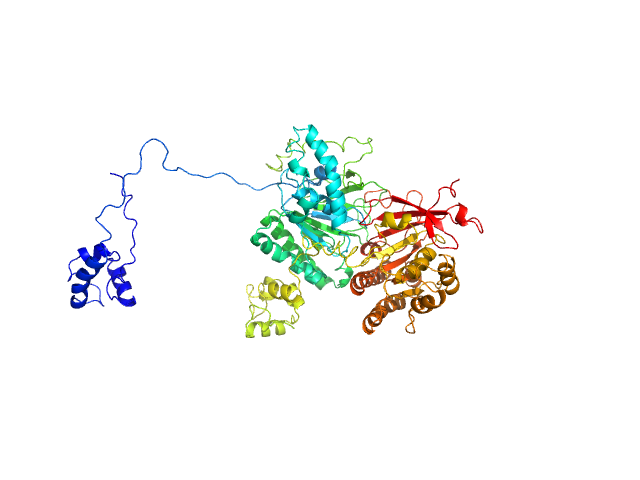
|
| Sample: |
Isoform 1 of DNA repair protein RAD51 homolog 3 monomer, 40 kDa Homo sapiens protein
DNA repair protein XRCC3 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 8, 100 mM NaCl, 2.5 mM ATP, 2.5 mM MgCl2, and 0.1 mM Na3VO4, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 May 5
|
RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles
Nature Communications 14(1) (2023)
Longo M, Roy S, Chen Y, Tomaszowski K, Arvai A, Pepper J, Boisvert R, Kunnimalaiyaan S, Keshvani C, Schild D, Bacolla A, Williams G, Tainer J, Schlacher K
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|

|
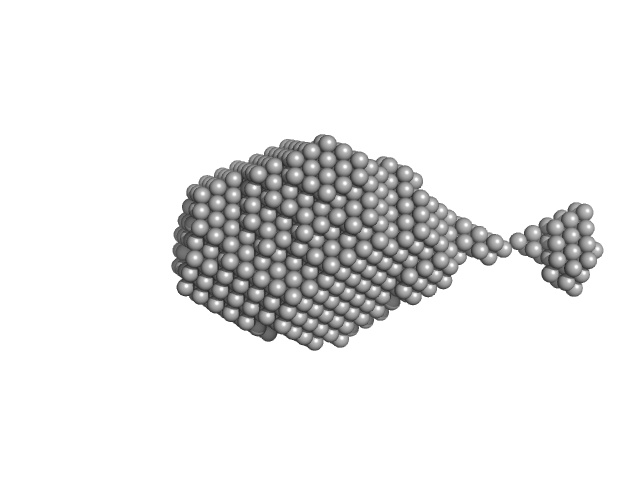
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
|
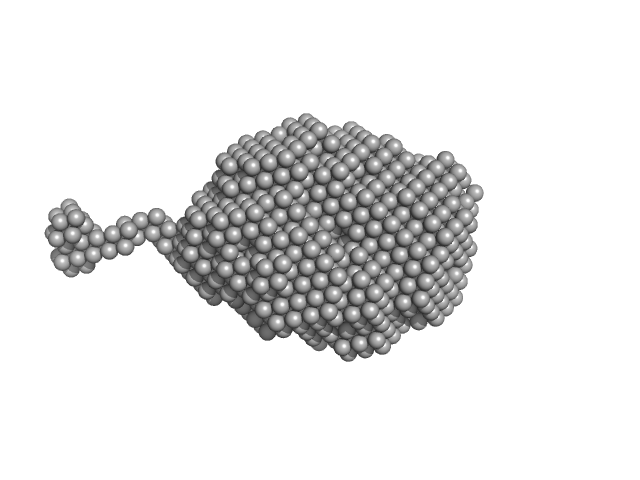
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|
|
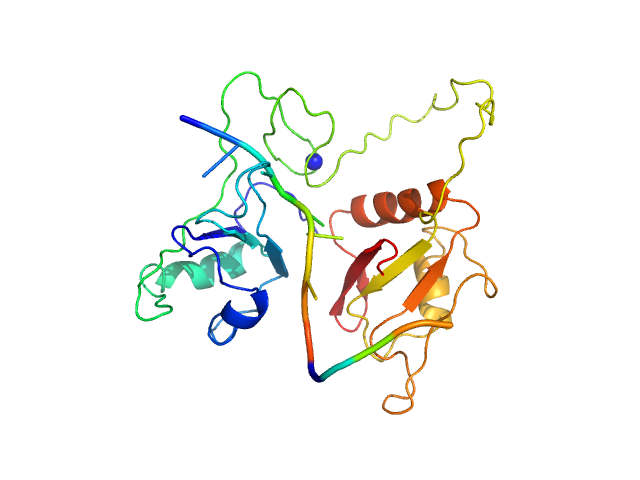
| Sample: |
RNA Binding Motif protein 5 (I107T, C191G) monomer, 26 kDa Homo sapiens protein
Caspase-2 derived RNA GGCU_12 monomer, 4 kDa RNA
|
| Buffer: |
20 mM MES, 100 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2017 Mar 1
|
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.
Nat Commun 14(1):4233 (2023)
Soni K, Jagtap PKA, Martínez-Lumbreras S, Bonnal S, Geerlof A, Stehle R, Simon B, Valcárcel J, Sattler M
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
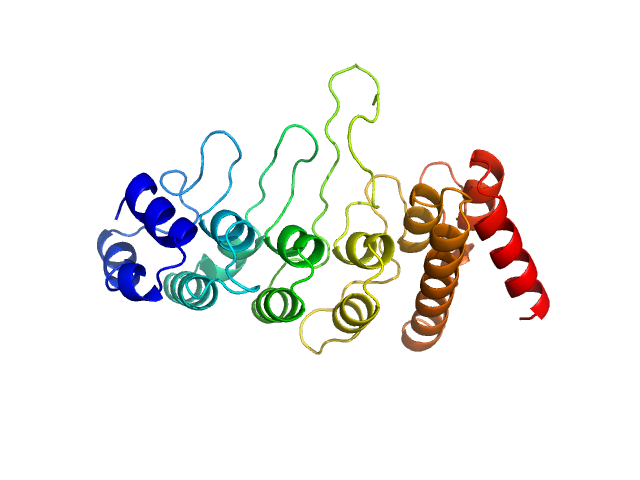
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 28 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Monooxygenase (M154I, A283T) monomer, 39 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jan 10
|
Tetracycline-modifying enzyme Sm
TetX from Stenotrophomonas maltophilia
Acta Crystallographica Section F Structural Biology Communications 79(7):180-192 (2023)
Malý M, Kolenko P, Stránský J, Švecová L, Dušková J, Koval' T, Skálová T, Trundová M, Adámková K, Černý J, Božíková P, Dohnálek J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
59 |
nm3 |
|
|