|
|
|
|
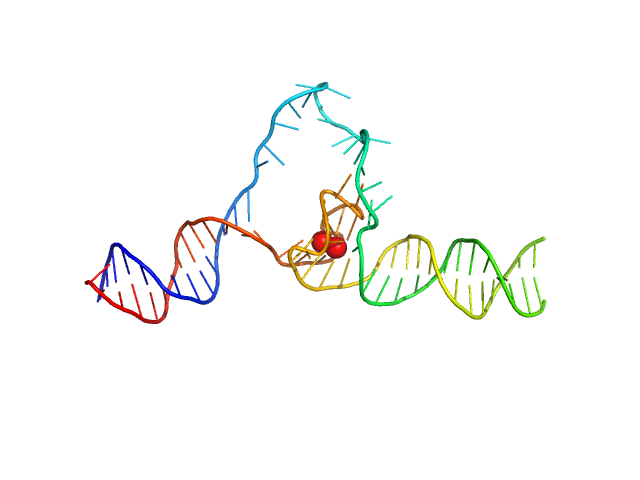
|
| Sample: |
Duplex-G4-duplex monomer, 29 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jun 16
|
Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 Å resolution with cryo-EM.
Nucleic Acids Res (2023)
Monsen RC, Chua EYD, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
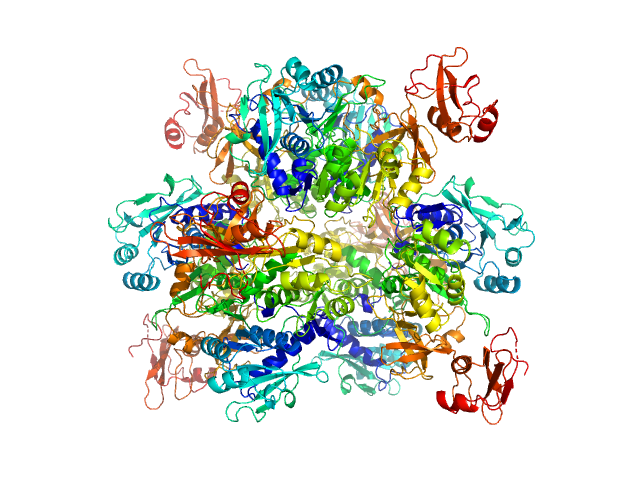
|
| Sample: |
Oxalate--CoA ligase, 363 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
565 |
nm3 |
|
|
|
|
|
|
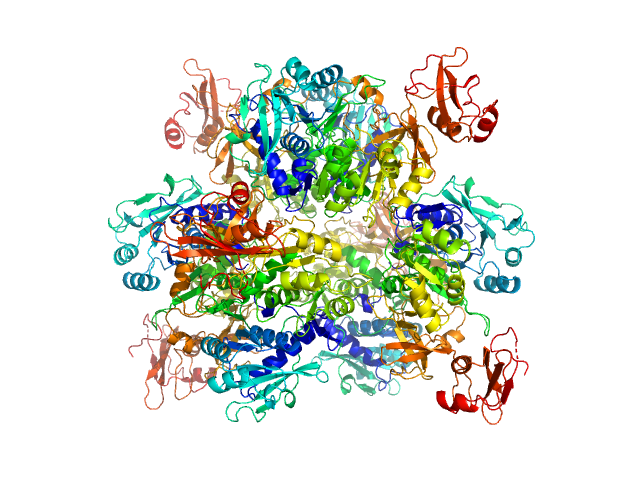
|
| Sample: |
Oxalate--CoA ligase, 363 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
432 |
nm3 |
|
|
|
|
|
|
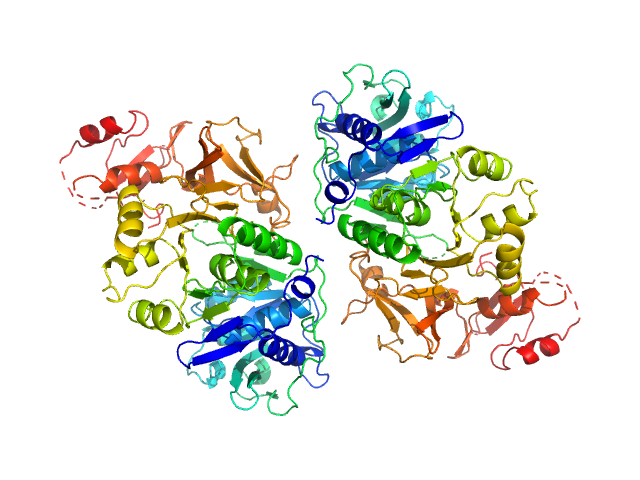
|
| Sample: |
Oxalate--CoA ligase (K352D) dimer, 121 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
|
|
|
|
|
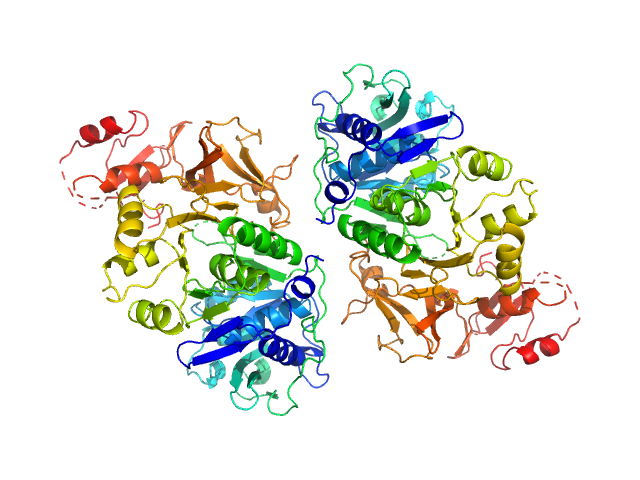
| Sample: |
Oxalate--CoA ligase (K352D) dimer, 121 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|
|
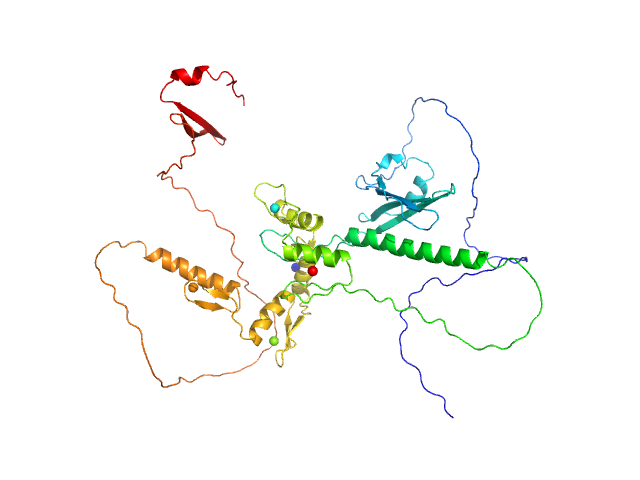
| Sample: |
Zinc finger protein 410 monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
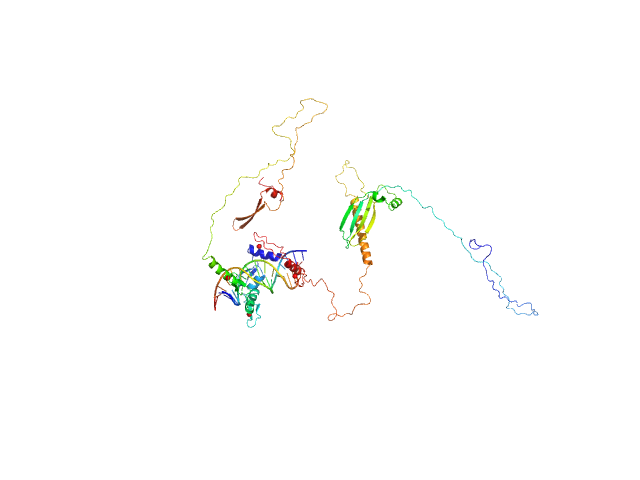
| Sample: |
Zinc finger protein 410 monomer, 52 kDa Homo sapiens protein
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
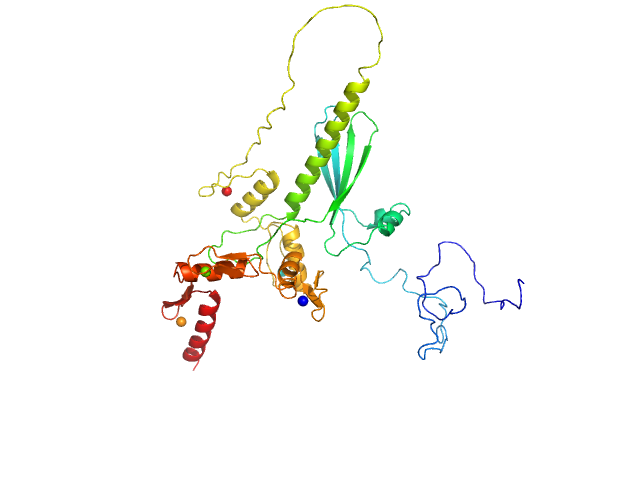
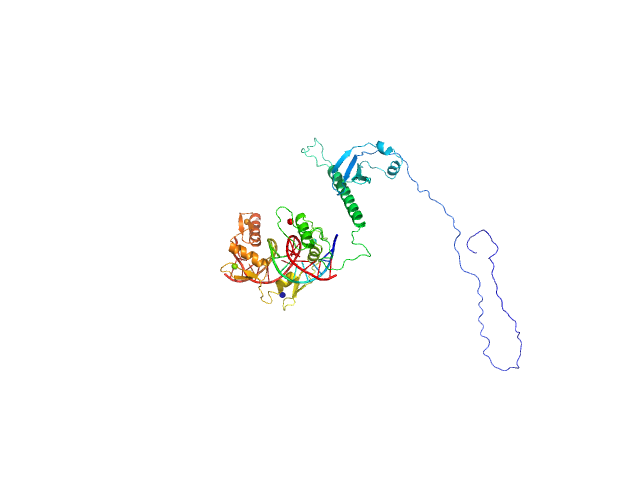
| Sample: |
Zinc finger protein 410 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
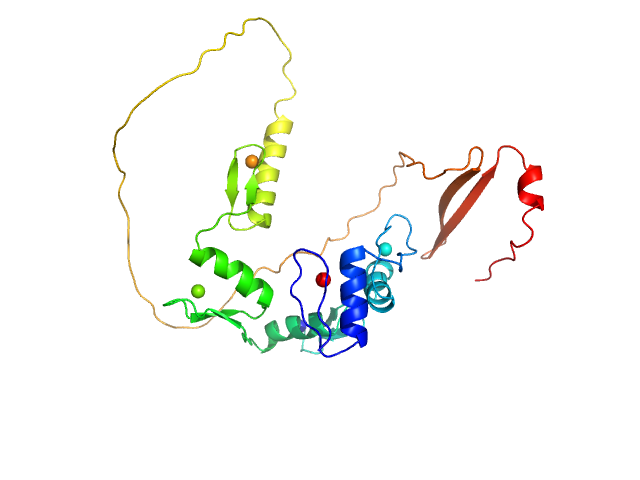
| Sample: |
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
Zinc finger protein 410 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Apr 25
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Zinc finger protein 410 monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|