|
|
|
|
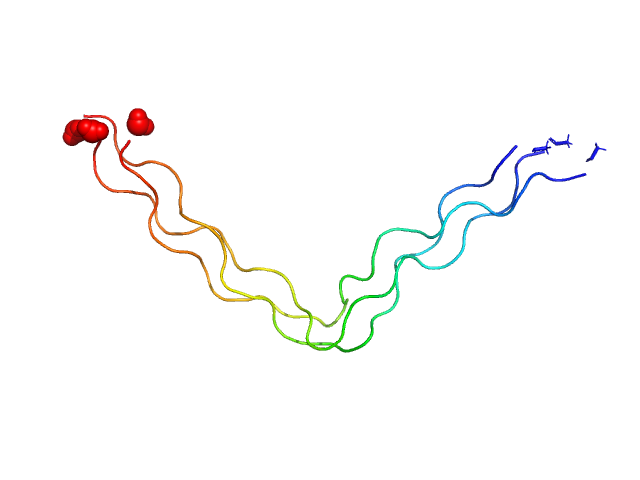
|
| Sample: |
Ac-(POG)4-QG-(POG)5-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ac-(POG)5-EOGQGLRG-(POG)3-NH2 trimer, 12 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
6 |
nm3 |
|
|
|
|
|
|
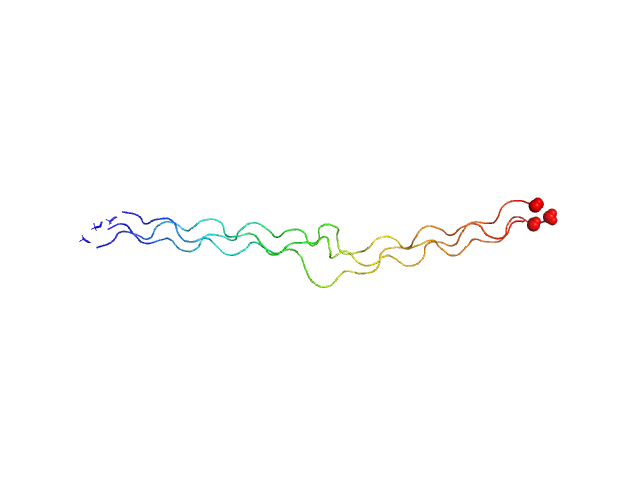
|
| Sample: |
Ac-(POG)5-EPGQGLRG-(POG)5-NH2 trimer, 14 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
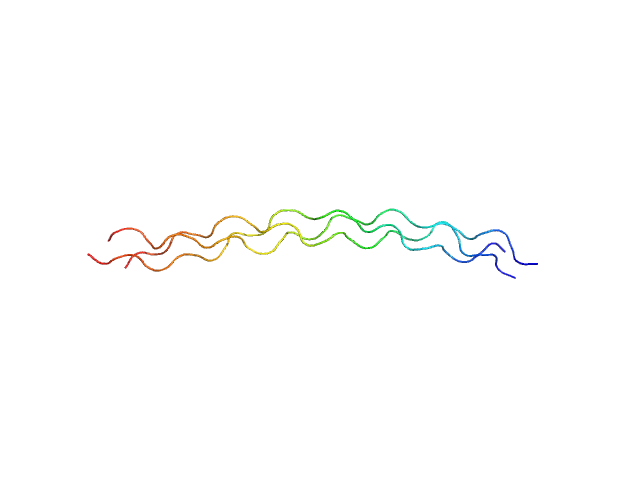
|
| Sample: |
Ac-(POG)10-NH2 trimer, 12 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
|
|
|
|
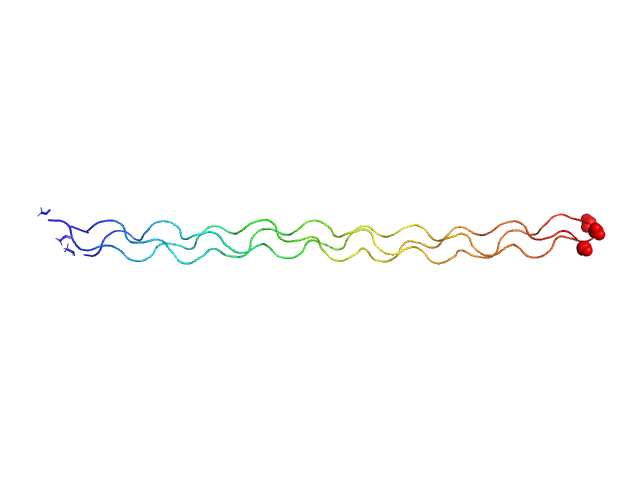
|
| Sample: |
Ac-(POG)13-NH2 trimer, 15 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
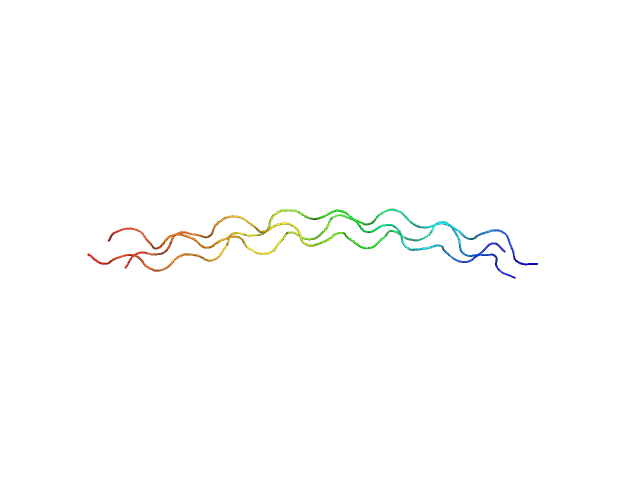
|
| Sample: |
[(Pro-Pro-Gly)10]3 trimer, 8 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ac-(POG)4-POA-(POG)5-NH2 trimer, 12 kDa protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ac-(POG)3-ITGARGLAG-(POG)4-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|
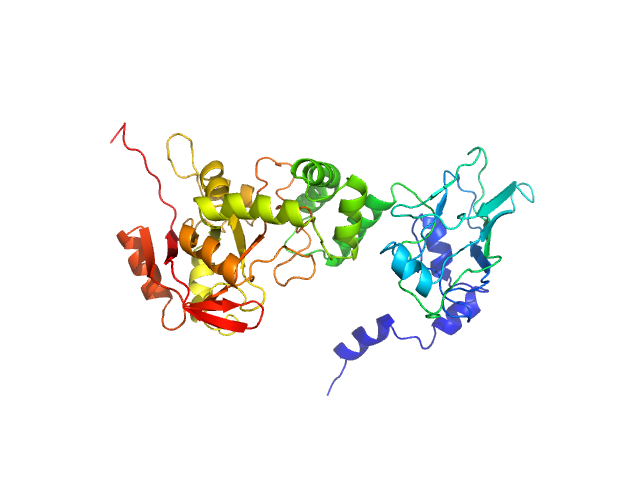
|
| Sample: |
NAD glycohydrolase monomer, 47 kDa Streptococcus pyogenes M1 … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Oct 16
|
Structural basis underlying the synergism of NADase and SLO during group A Streptococcus infection.
Commun Biol 6(1):124 (2023)
Tsai WJ, Lai YH, Shi YA, Hammel M, Duff AP, Whitten AE, Wilde KL, Wu CM, Knott R, Jeng US, Kang CY, Hsu CY, Wu JL, Tsai PJ, Chiang-Ni C, Wu JJ, Lin YS, Liu CC, Senda T, Wang S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
103.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
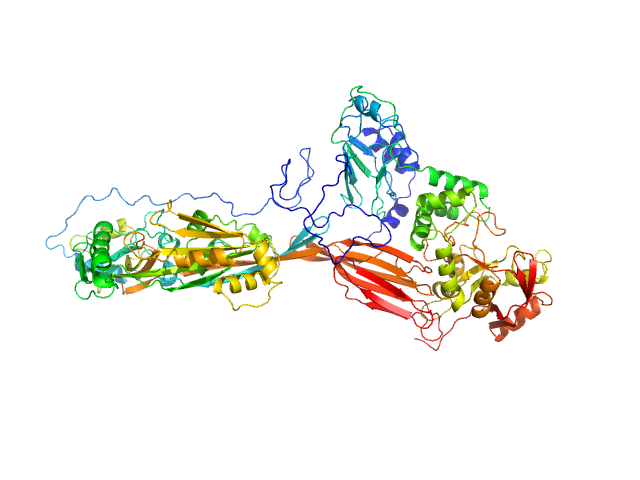
|
| Sample: |
NAD glycohydrolase monomer, 47 kDa Streptococcus pyogenes M1 … protein
Streptolysin O (T66M) monomer, 63 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Oct 16
|
Structural basis underlying the synergism of NADase and SLO during group A Streptococcus infection.
Commun Biol 6(1):124 (2023)
Tsai WJ, Lai YH, Shi YA, Hammel M, Duff AP, Whitten AE, Wilde KL, Wu CM, Knott R, Jeng US, Kang CY, Hsu CY, Wu JL, Tsai PJ, Chiang-Ni C, Wu JJ, Lin YS, Liu CC, Senda T, Wang S
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
125 |
nm3 |
|
|