|
|
|
|
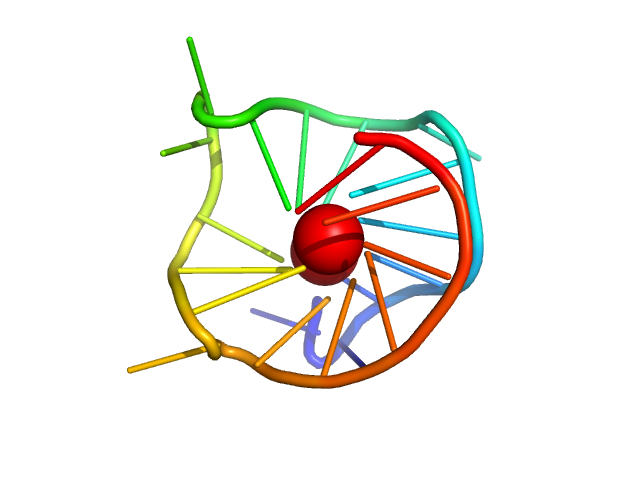
|
| Sample: |
CMyc promoter modified GQ parallel monomer, 7 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 11
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.1 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|
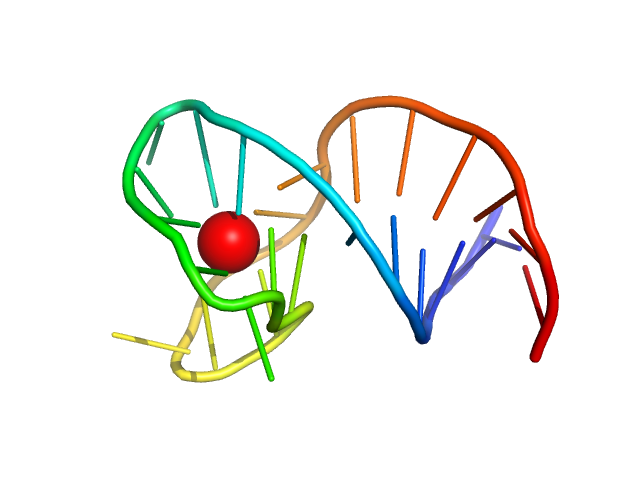
|
| Sample: |
Two-tetrad antiparallel GQ with duplex junction monomer, 10 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 3
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
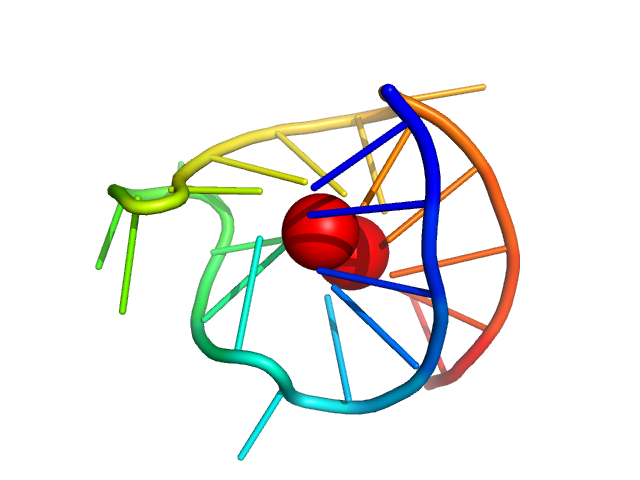
|
| Sample: |
VEGF promoter GQ parallel monomer, 7 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 20
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.3 |
nm |
| Dmax |
3.8 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
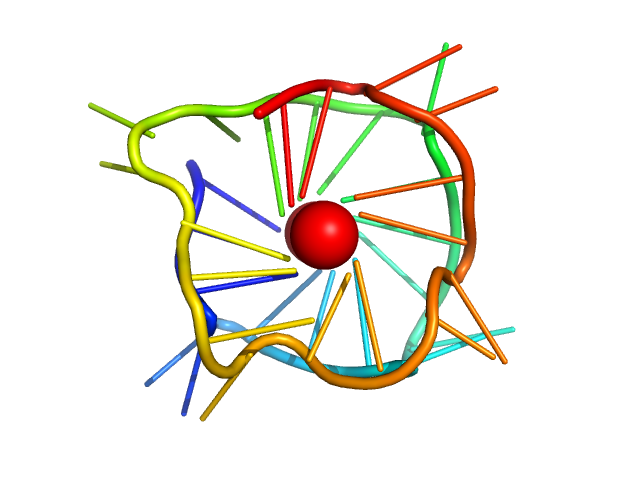
|
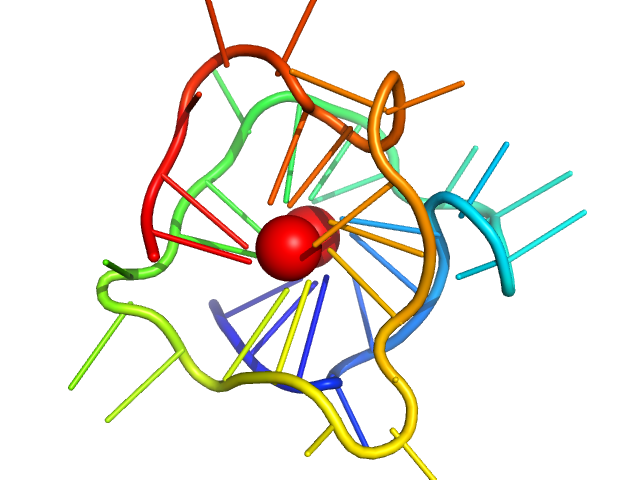
| Sample: |
CMyc promoter 8-tract G-quadruplex monomer, 11 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 19
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
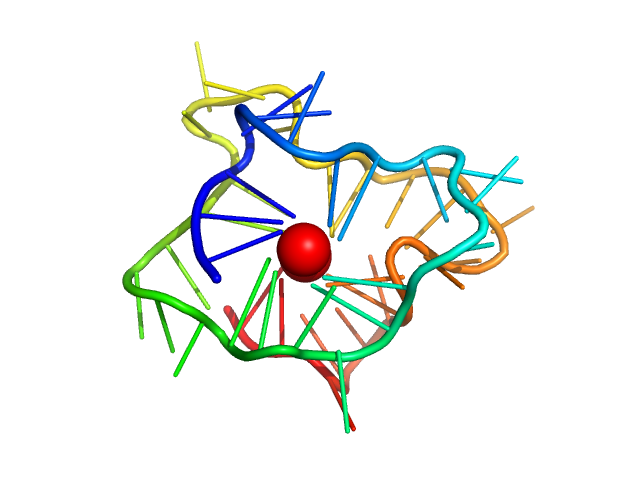
| Sample: |
CKit promoter 8-tract G-quadruplex monomer, 12 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 19
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.7 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
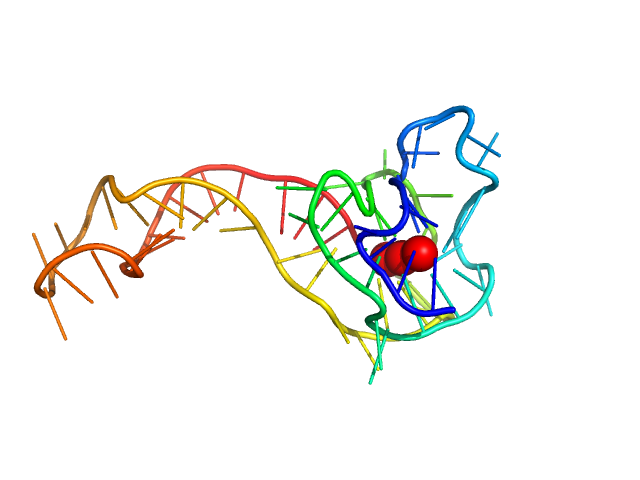
| Sample: |
KRas promoter 8-tract G-quadruplex monomer, 14 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 19
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CKit promoter 12-tract G-quadruplex monomer, 20 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 22
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
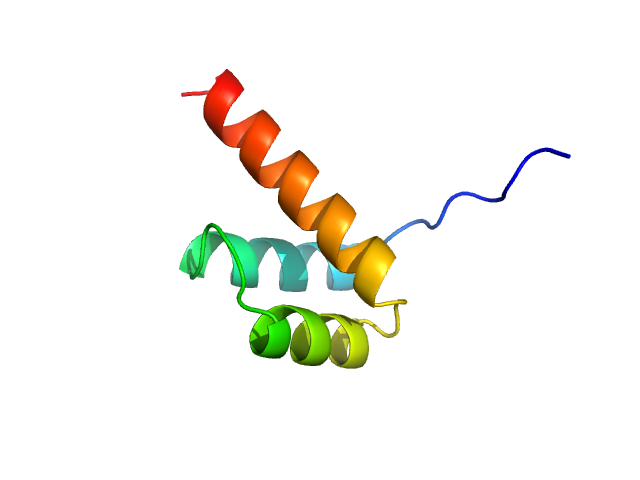
| Sample: |
Cone-rod homeobox protein monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate,100 mM NaCl, and 5 mM imidazole, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Jun 15
|
Structural and functional analysis of the human Cone-rod homeobox transcription factor.
Proteins (2022)
Clanor PB, Buchholz C, Hayes JE, Friedman MA, White AM, Enke RA, Berndsen CE
|
| RgGuinier |
1.2 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
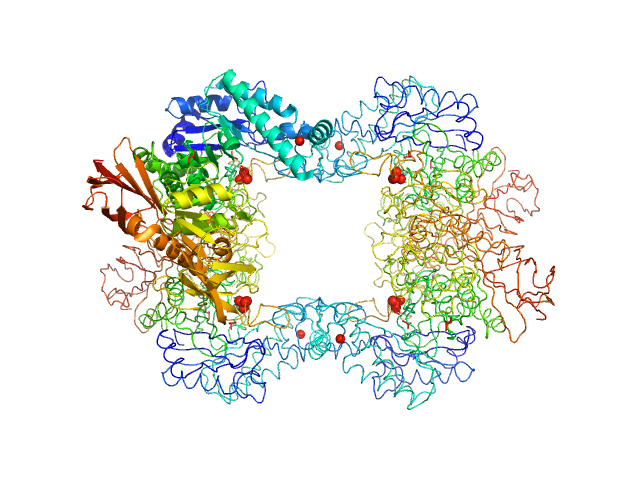
|
| Sample: |
Alpha-L-fucosidase tetramer, 297 kDa Paenibacillus thiaminolyticus (Bacillus … protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2019 Jul 16
|
The first structure–function study of GH151 α‐ l
‐fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
The FEBS Journal (2022)
Koval'ová T, Kovaľ T, Stránský J, Kolenko P, Dušková J, Švecová L, Vodičková P, Spiwok V, Benešová E, Lipovová P, Dohnálek J
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
418 |
nm3 |
|
|
|
|
|
|
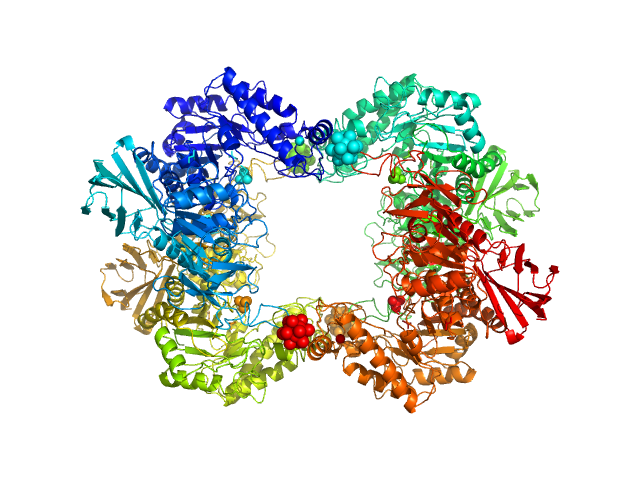
|
| Sample: |
Alpha-L-fucosidase H503A tetramer, 297 kDa Paenibacillus thiaminolyticus (Bacillus … protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2019 Jul 16
|
The first structure–function study of GH151 α‐ l
‐fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
The FEBS Journal (2022)
Koval'ová T, Kovaľ T, Stránský J, Kolenko P, Dušková J, Švecová L, Vodičková P, Spiwok V, Benešová E, Lipovová P, Dohnálek J
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
433 |
nm3 |
|
|