|
|
|
|
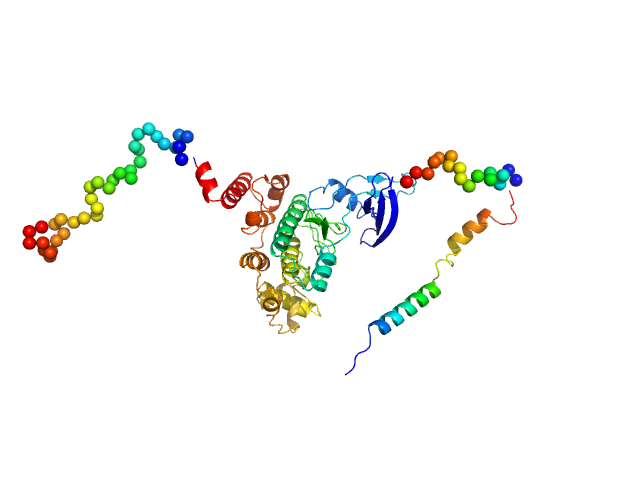
|
| Sample: |
Glycogen synthase kinase 3 monomer, 52 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 19
|
N-terminal phosphorylation regulates the activity of glycogen synthase kinase 3 from Plasmodium falciparum.
Biochem J 479(3):337-356 (2022)
Pazicky S, Alder A, Mertens H, Svergun D, Gilberger T, Löw C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
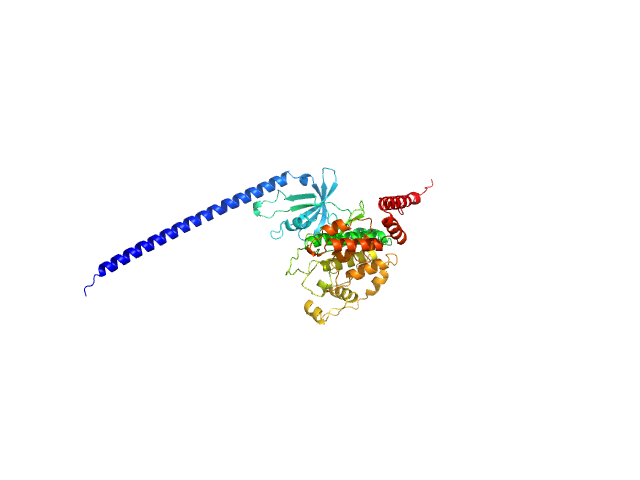
|
| Sample: |
Glycogen synthase kinase 3 monomer, 52 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 19
|
N-terminal phosphorylation regulates the activity of glycogen synthase kinase 3 from Plasmodium falciparum.
Biochem J 479(3):337-356 (2022)
Pazicky S, Alder A, Mertens H, Svergun D, Gilberger T, Löw C
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hemolysin, plasmid (Hemolysin A) dimer, 220 kDa Escherichia coli UTI89 protein
|
| Buffer: |
100 mM HEPES pH 8.0, 250 mM NaCl, 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 6
|
Identity Determinants of the Translocation Signal for a Type 1 Secretion System
Frontiers in Physiology 12 (2022)
Spitz O, Erenburg I, Kanonenberg K, Peherstorfer S, Lenders M, Reiners J, Ma M, Luisi B, Smits S, Schmitt L
|
| RgGuinier |
6.7 |
nm |
| Dmax |
25.3 |
nm |
| VolumePorod |
346 |
nm3 |
|
|
|
|
|
|
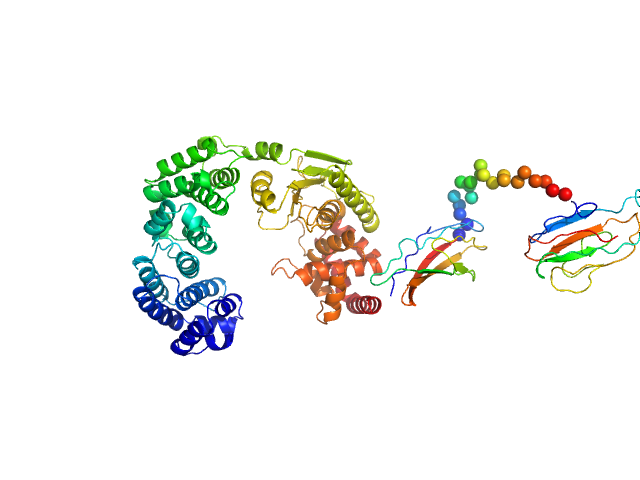
|
| Sample: |
Vibrio collagenase VhaC monomer, 90 kDa Vibrio harveyi protein
|
| Buffer: |
10 mM Tris-HCl, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jan 1
|
Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun 13(1):566 (2022)
Wang Y, Wang P, Cao HY, Ding HT, Su HN, Liu SC, Liu G, Zhang X, Li CY, Peng M, Li F, Li S, Chen Y, Chen XL, Zhang YZ
|
| RgGuinier |
4.3 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
148 |
nm3 |
|
|
|
|
|
|
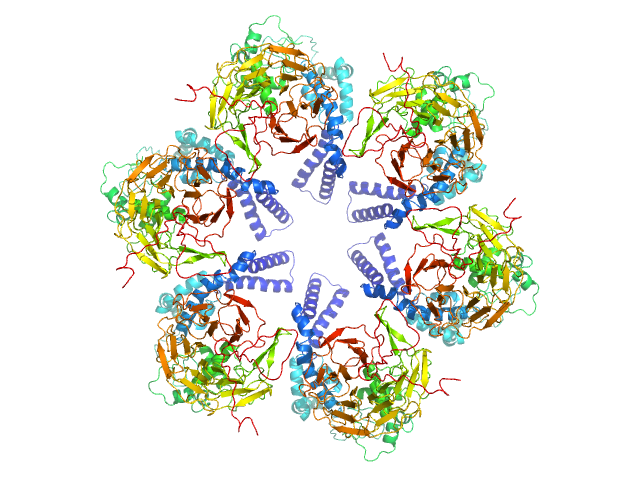
|
| Sample: |
Kelch protein K13 hexamer, 396 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2020 Jul 25
|
Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: a SAXS data driven modeling study.
FEBS J (2022)
Goel N, Dhiman K, Kalidas N, Mukhopadhyay A, Ashish F, Bhattacharjee S
|
| RgGuinier |
6.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
1630 |
nm3 |
|
|
|
|
|
|
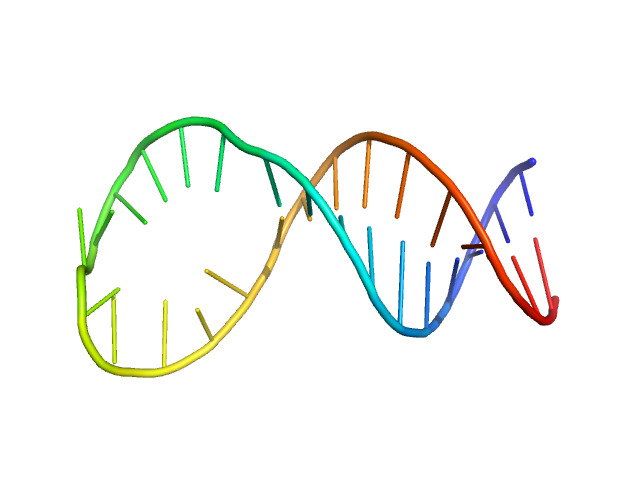
|
| Sample: |
SsDNA aptamer Apt31 specific to the receptor-binding domain of SARS-CoV-2 monomer, 10 kDa Artificially synthesized DNA
|
| Buffer: |
Tris-HCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2020 Dec 18
|
Structure and Interaction Based Design of Anti‐SARS‐CoV‐2 Aptamers
Chemistry – A European Journal (2022)
Mironov V, Shchugoreva I, Artyushenko P, Morozov D, Borbone N, Oliviero G, Zamay T, Moryachkov R, Kolovskaya O, Lukyanenko K, Song Y, Merkuleva I, Zabluda V, Peters G, Koroleva L, Veprintsev D, Glazyrin Y, Volosnikova E, Belenkaya S, Esina T, Isaeva A, Nesmeyanova V, Shanshin D, Berlina A, Komova N, Svetlichnyi V, Silnikov V, Shcherbakov D, Zamay G, Zamay S, Smolyarova T, Tikhonova E, Chen K, Jeng U, Condorelli G, de Franciscis V, Groenhof G, Yang C, Moskovsky A, Fedorov D, Tomilin F, Tan W, Alexeev Y, Berezovski M, Kichkailo A
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Lectin nano-block WA20-SL-ACG dimer, 60 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Jun 20
|
Self-Assembling Lectin Nano-Block Oligomers Enhance Binding Avidity to Glycans
International Journal of Molecular Sciences 23(2):676 (2022)
Irumagawa S, Hiemori K, Saito S, Tateno H, Arai R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|
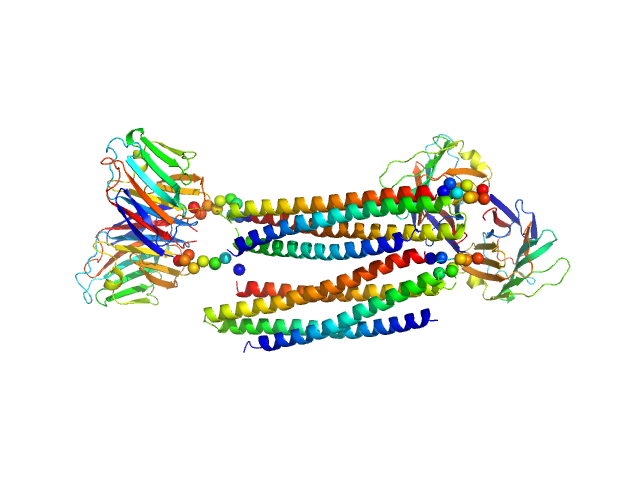
|
| Sample: |
Lectin nano-block WA20-SL-ACG tetramer, 121 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Jun 20
|
Self-Assembling Lectin Nano-Block Oligomers Enhance Binding Avidity to Glycans
International Journal of Molecular Sciences 23(2):676 (2022)
Irumagawa S, Hiemori K, Saito S, Tateno H, Arai R
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.6 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
|
|
|
|
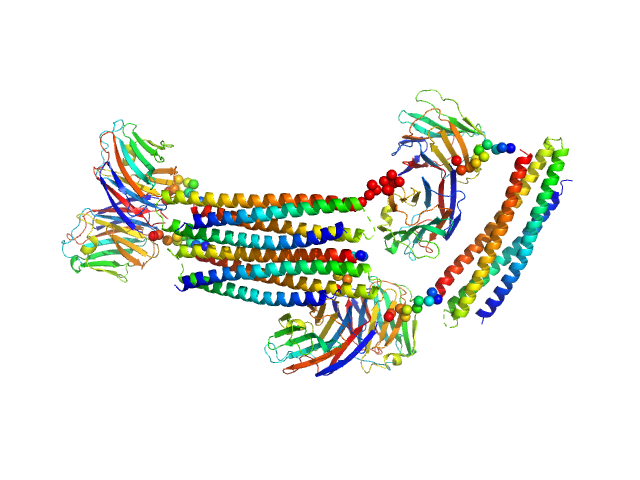
|
| Sample: |
Lectin nano-block WA20-SL-ACG hexamer, 181 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Jun 20
|
Self-Assembling Lectin Nano-Block Oligomers Enhance Binding Avidity to Glycans
International Journal of Molecular Sciences 23(2):676 (2022)
Irumagawa S, Hiemori K, Saito S, Tateno H, Arai R
|
| RgGuinier |
5.4 |
nm |
| Dmax |
24.2 |
nm |
| VolumePorod |
313 |
nm3 |
|
|
|
|
|
|
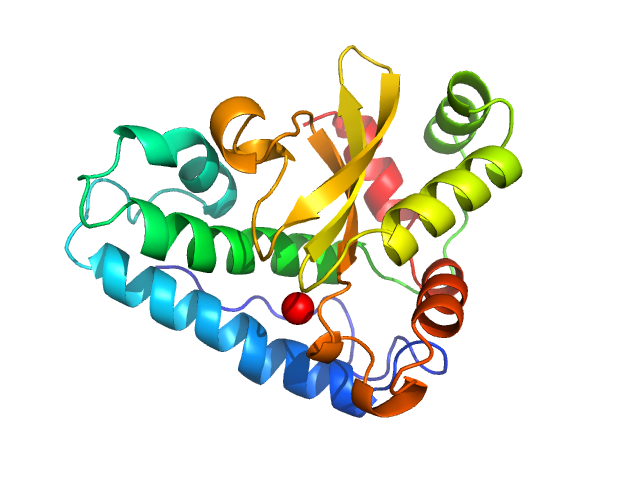
|
| Sample: |
Superoxide dismutase [Mn], 23 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 1
|
Protein quaternary structures in solution are a mixture of multiple forms
Chemical Science 13(39):11680-11695 (2022)
Marciano S, Dey D, Listov D, Fleishman S, Sonn-Segev A, Mertens H, Busch F, Kim Y, Harvey S, Wysocki V, Schreiber G
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
54 |
nm3 |
|
|