|
|
|
|
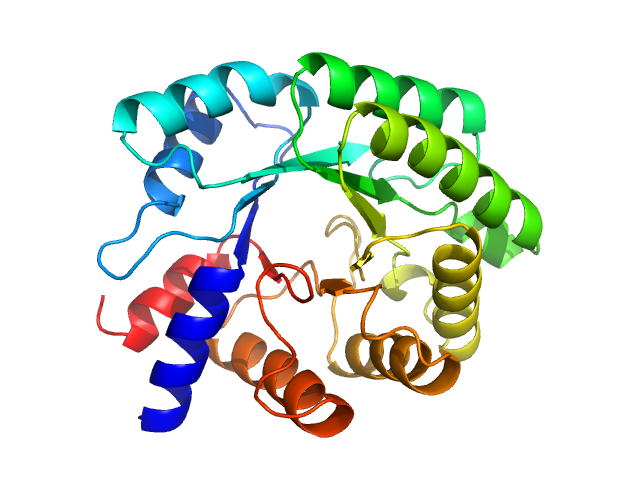
|
| Sample: |
Deoxyribose-phosphate aldolase, 28 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 1
|
Protein quaternary structures in solution are a mixture of multiple forms
Chemical Science 13(39):11680-11695 (2022)
Marciano S, Dey D, Listov D, Fleishman S, Sonn-Segev A, Mertens H, Busch F, Kim Y, Harvey S, Wysocki V, Schreiber G
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
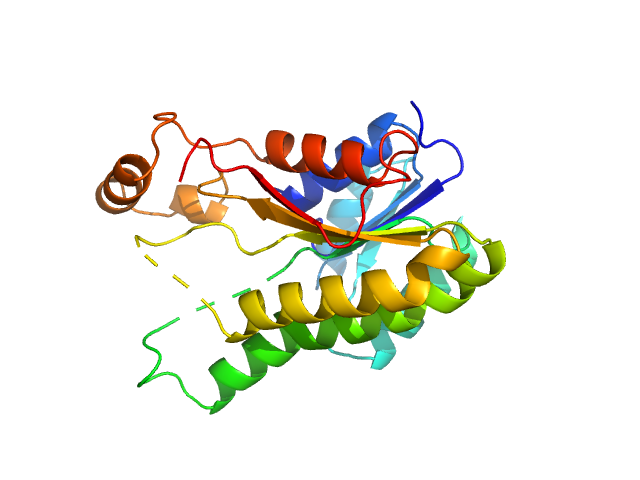
|
| Sample: |
3-oxoacyl-[acyl-carrier-protein] reductase FabG, 26 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 1
|
Protein quaternary structures in solution are a mixture of multiple forms
Chemical Science 13(39):11680-11695 (2022)
Marciano S, Dey D, Listov D, Fleishman S, Sonn-Segev A, Mertens H, Busch F, Kim Y, Harvey S, Wysocki V, Schreiber G
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
|
|
|
|
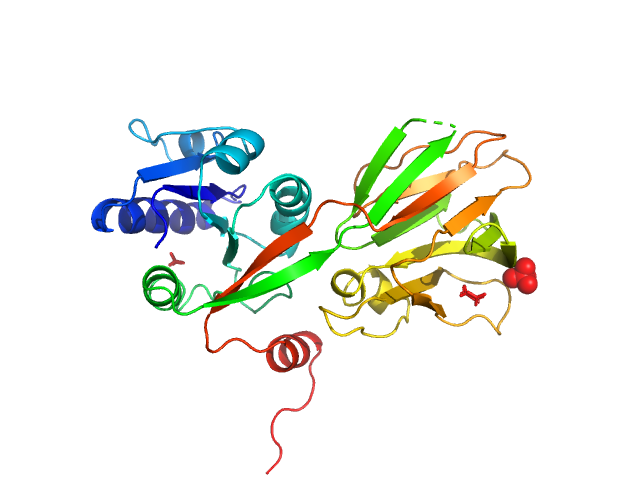
|
| Sample: |
NAD kinase, 33 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM HEPES, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 2
|
Protein quaternary structures in solution are a mixture of multiple forms
Chemical Science 13(39):11680-11695 (2022)
Marciano S, Dey D, Listov D, Fleishman S, Sonn-Segev A, Mertens H, Busch F, Kim Y, Harvey S, Wysocki V, Schreiber G
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
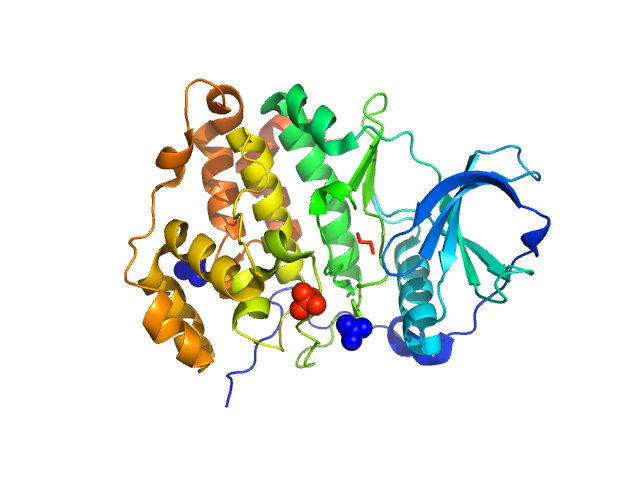
|
| Sample: |
Casein kinase II subunit alpha monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 500 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Oct 10
|
Mechanism of CK2 Inhibition by a Ruthenium-Based Polyoxometalate.
Front Mol Biosci 9:906390 (2022)
Fabbian S, Giachin G, Bellanda M, Borgo C, Ruzzene M, Spuri G, Campofelice A, Veneziano L, Bonchio M, Carraro M, Battistutta R
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
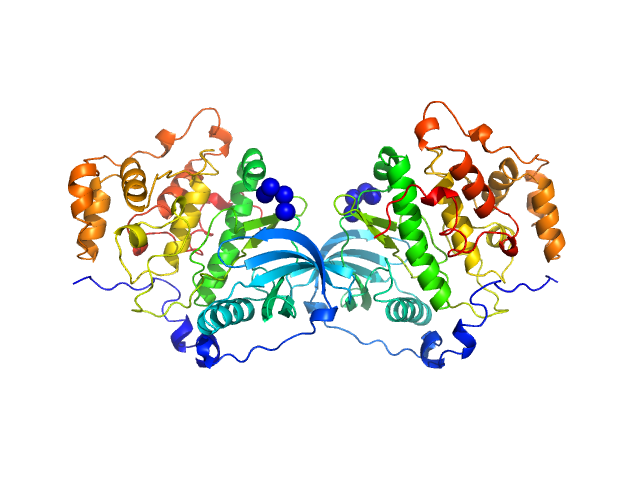
| Sample: |
Casein kinase II subunit alpha dimer, 78 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 500 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Oct 10
|
Mechanism of CK2 Inhibition by a Ruthenium-Based Polyoxometalate.
Front Mol Biosci 9:906390 (2022)
Fabbian S, Giachin G, Bellanda M, Borgo C, Ruzzene M, Spuri G, Campofelice A, Veneziano L, Bonchio M, Carraro M, Battistutta R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
|
|
|
|
|
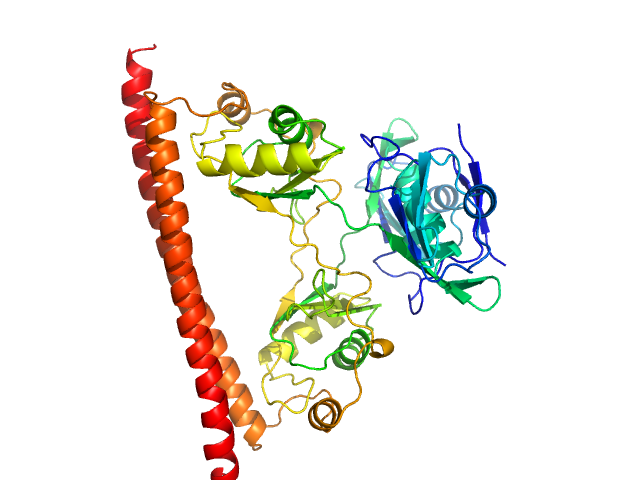
| Sample: |
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-Cl (pH 7.5), 250 mM KCl, 50 mM L-proline, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural basis of dimerization and nucleic acid binding of human DBHS proteins NONO and PSPC1.
Nucleic Acids Res (2021)
Knott GJ, Chong YS, Passon DM, Liang XH, Deplazes E, Conte MR, Marshall AC, Lee M, Fox AH, Bond CS
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
|
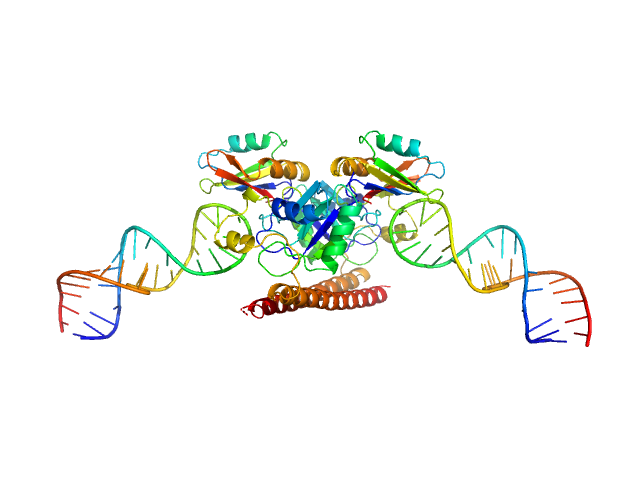
| Sample: |
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
5-10-5 gapmer phosphorothioate antisense oligonucleotide tetramer tetramer, 28 kDa
|
| Buffer: |
20 mM Tris-Cl (pH 7.5), 250 mM KCl, 50 mM L-proline, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural basis of dimerization and nucleic acid binding of human DBHS proteins NONO and PSPC1.
Nucleic Acids Res (2021)
Knott GJ, Chong YS, Passon DM, Liang XH, Deplazes E, Conte MR, Marshall AC, Lee M, Fox AH, Bond CS
|
| RgGuinier |
3.9 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
153 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Serotransferrin monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 20 mM NaHCO3, 50 mM NaCl, (APO Buffer), pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin
International Journal of Molecular Sciences 22(24):13392 (2021)
Campos-Escamilla C, Siliqi D, Gonzalez-Ramirez L, Lopez-Sanchez C, Gavira J, Moreno A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
|
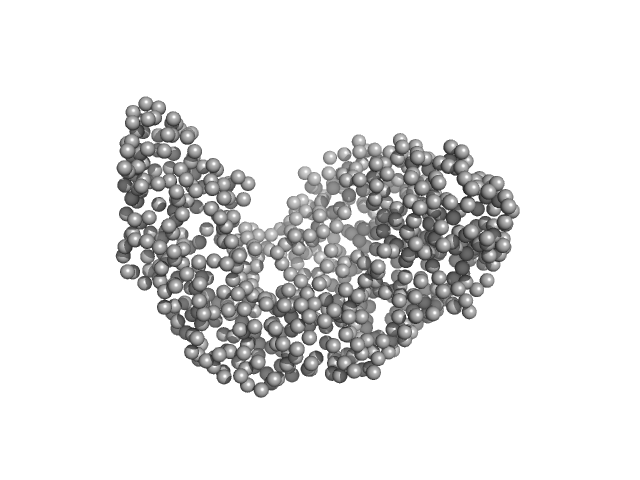
| Sample: |
Serotransferrin monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 20 mM NaHCO3, 50 mM NaCl (APO Buffer), pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin
International Journal of Molecular Sciences 22(24):13392 (2021)
Campos-Escamilla C, Siliqi D, Gonzalez-Ramirez L, Lopez-Sanchez C, Gavira J, Moreno A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
|
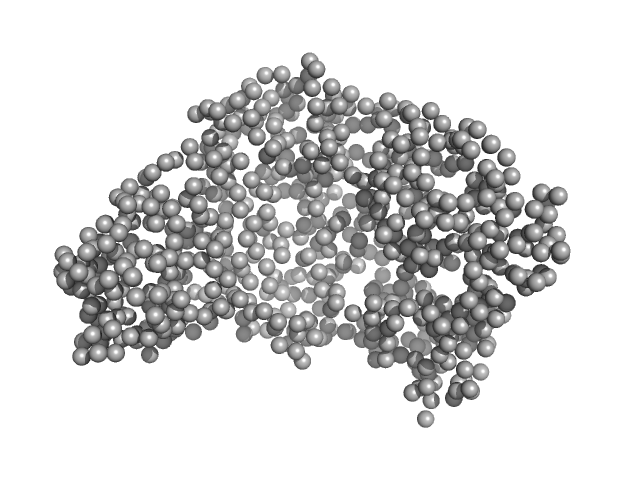
| Sample: |
Serotransferrin monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 20 mM NaHCO3, 50 mM NaCl (Buffer APO-Tf-1), pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin
International Journal of Molecular Sciences 22(24):13392 (2021)
Campos-Escamilla C, Siliqi D, Gonzalez-Ramirez L, Lopez-Sanchez C, Gavira J, Moreno A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
106 |
nm3 |
|
|