|
|
|
|

|
| Sample: |
Aquifex aeolicus McoA metaloxidase ∆328-352 (MCoA∆328-352) monomer, 53 kDa Aquifex aeolicus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 13
|
The Methionine-Rich Loop of Multicopper Oxidase McoA follows Open-To-Close Transitions with a Role in Enzyme Catalysis
ACS Catalysis (2020)
Borges P, Brissos V, Hernandez G, Masgrau L, Lucas M, Monza E, Frazão C, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aquifex aeolicus McoA metaloxidase ∆337-346 monomer, 54 kDa Aquifex aeolicus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Dec 4
|
The Methionine-Rich Loop of Multicopper Oxidase McoA follows Open-To-Close Transitions with a Role in Enzyme Catalysis
ACS Catalysis (2020)
Borges P, Brissos V, Hernandez G, Masgrau L, Lucas M, Monza E, Frazão C, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aquifex aeolicus McoA metaloxidase monomer, 55 kDa Aquifex aeolicus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 15
|
The Methionine-Rich Loop of Multicopper Oxidase McoA follows Open-To-Close Transitions with a Role in Enzyme Catalysis
ACS Catalysis (2020)
Borges P, Brissos V, Hernandez G, Masgrau L, Lucas M, Monza E, Frazão C, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
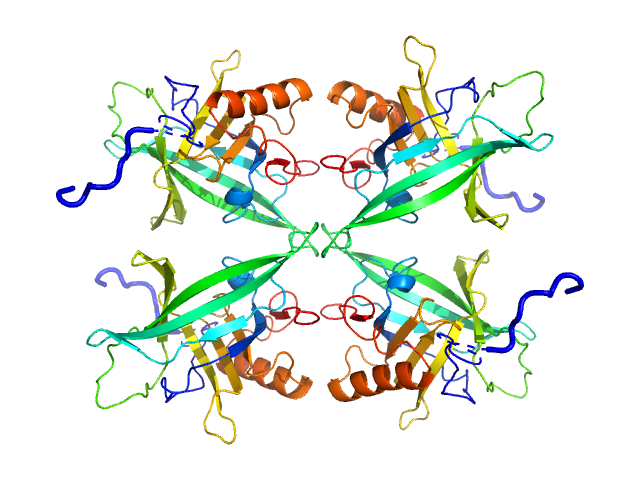
|
| Sample: |
Plasmodium falciparum Lipocalin tetramer, 89 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM Tris pH7.5, 150 mM NaCl, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum
Cell Reports 31(12):107817 (2020)
Burda P, Crosskey T, Lauk K, Zurborg A, Söhnchen C, Liffner B, Wilcke L, Pietsch E, Strauss J, Jeffries C, Svergun D, Wilson D, Wilmanns M, Gilberger T
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
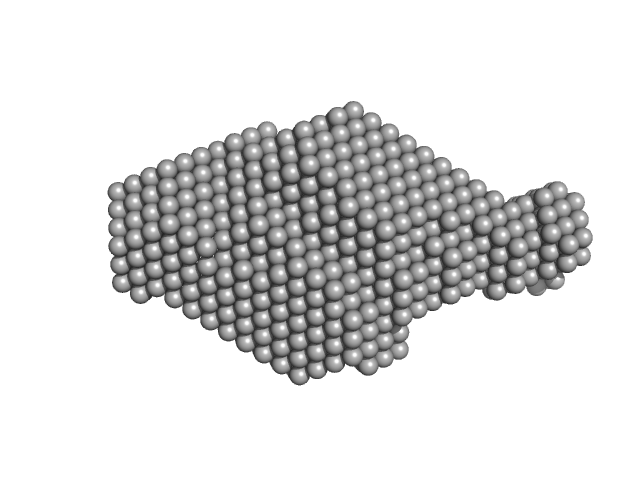
|
| Sample: |
Tegument protein UL7 monomer, 34 kDa Human alphaherpesvirus 1 … protein
Tegument protein UL51 dimer, 30 kDa Human alphaherpesvirus 1 … protein
|
| Buffer: |
20 mM tris, 200 mM NaCl, 3% (v/v) glycerol, 0.25 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 16
|
Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex.
Elife 9 (2020)
Butt BG, Owen DJ, Jeffries CM, Ivanova L, Hill CH, Houghton JW, Ahmed MF, Antrobus R, Svergun DI, Welch JJ, Crump CM, Graham SC
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|
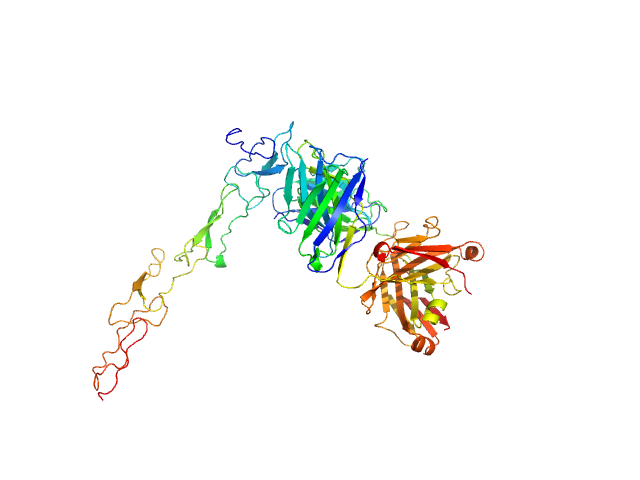
|
| Sample: |
Tumor necrosis factor receptor superfamily member 5 monomer, 19 kDa Homo sapiens protein
Human 341G2 F(ab) monomer, 73 kDa Homo sapiens protein
|
| Buffer: |
Phosphate-buffered saline, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
Isotype Switching Converts Anti-CD40 Antagonism to Agonism to Elicit Potent Antitumor Activity
Cancer Cell (2020)
Yu X, Chan H, Fisher H, Penfold C, Kim J, Inzhelevskaya T, Mockridge C, French R, Duriez P, Douglas L, English V, Verbeek J, White A, Tews I, Glennie M, Cragg M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
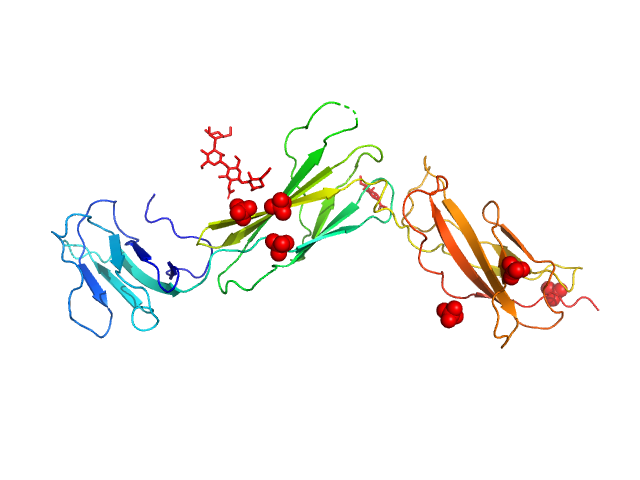
|
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
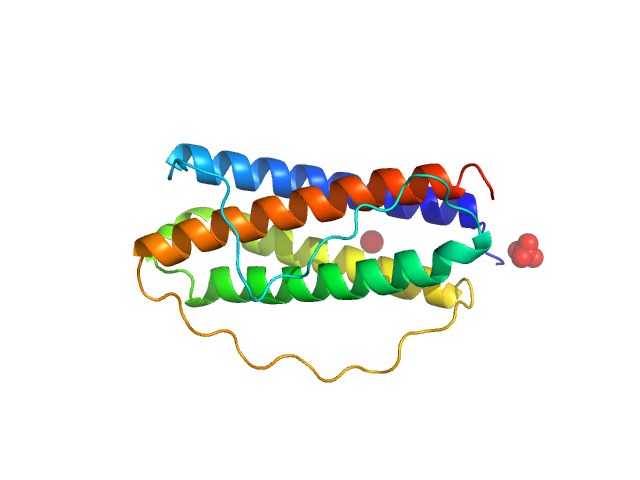
| Sample: |
Interleukin 11 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
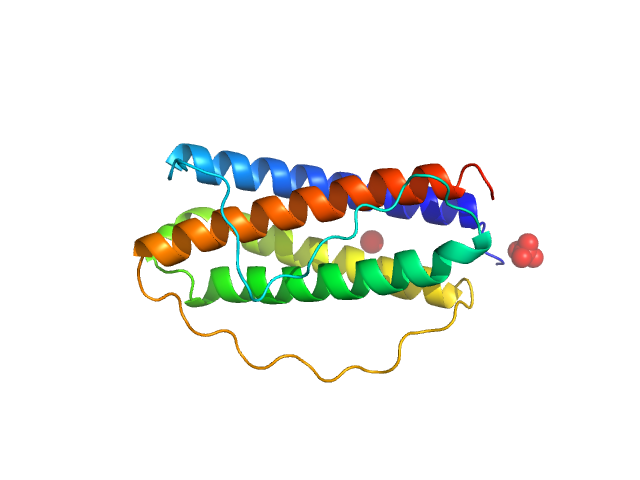
| Sample: |
Interleukin 11 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
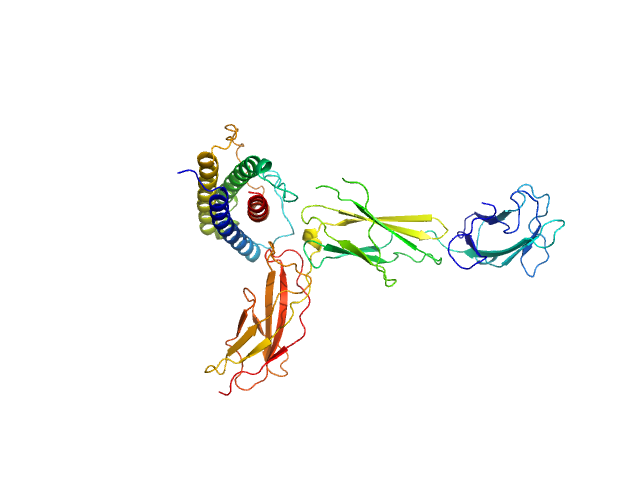
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
Interleukin 11 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
84 |
nm3 |
|
|