|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E34K dimer, 19 kDa Escherichia coli protein
|
| Buffer: |
10 mM sodium acetate, 50 mM NaCl, pH: 4.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 2
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E34K dimer, 19 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 50 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 2
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E34K, 100 kDa Escherichia coli protein
|
| Buffer: |
10mM Bis-Tris, 50 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 2
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
7.3 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E34K, 100 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 2
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
7.1 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Liver Receptor Homolog-1 monomer, 64 kDa Homo sapiens protein
Peroxisome Proliferator-Activated Receptor Gamma Coactivator-1 Alpha monomer, 2 kDa Homo sapiens protein
CYP7A1 Promoter Forward monomer, 4 kDa Homo sapiens DNA
CYP7A1 Promoter Reverse monomer, 4 kDa Homo sapiens DNA
|
| Buffer: |
20 mM TRIS, 150 mM NaCl, 2% v/v glycerol, 0.5 mM CHAPS, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 26
|
Integrated Structural Modeling of Full-Length LRH-1 Reveals Inter-domain Interactions Contribute to Receptor Structure and Function.
Structure (2020)
Seacrist CD, Kuenze G, Hoffmann RM, Moeller BE, Burke JE, Meiler J, Blind RD
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
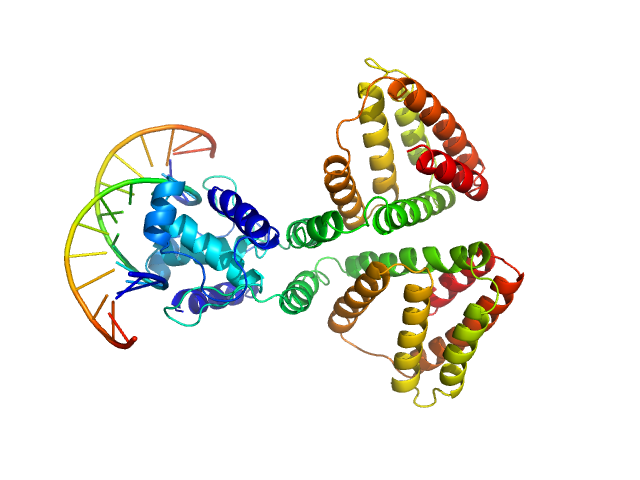
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
(GGTATA)2 repeat DNA monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
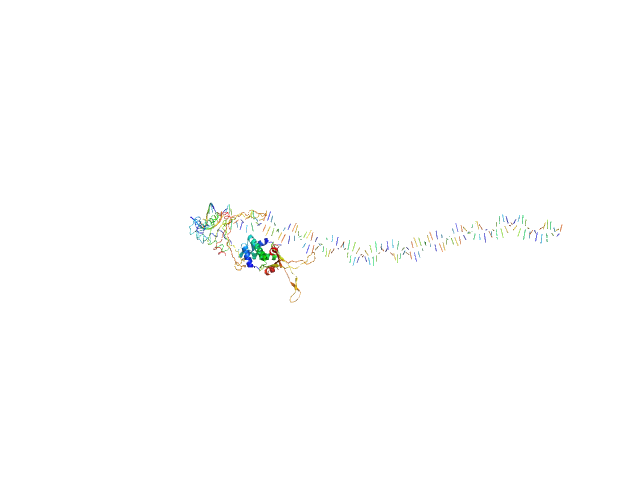
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E38K/V42L double mutant tetramer, 38 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Apr 23
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling (supplementary)
Soumya G Remesh
|
| RgGuinier |
5.7 |
nm |
| Dmax |
31.3 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
|
|
|
|
|
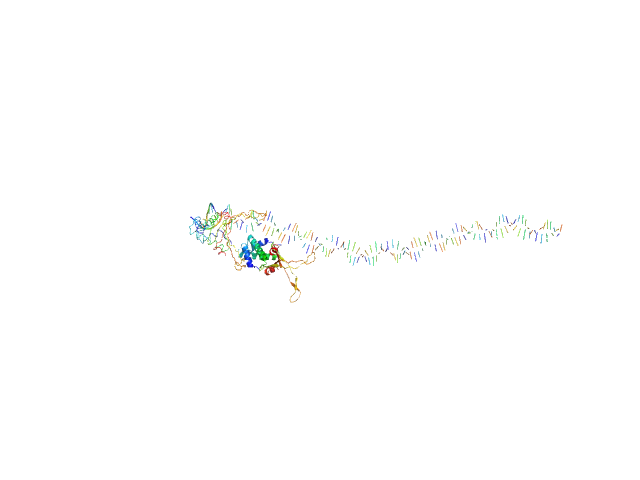
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E38K/V42L double mutant tetramer, 38 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Apr 23
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling (supplementary)
Soumya G Remesh
|
| RgGuinier |
5.7 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
195 |
nm3 |
|
|
|
|
|
|
|
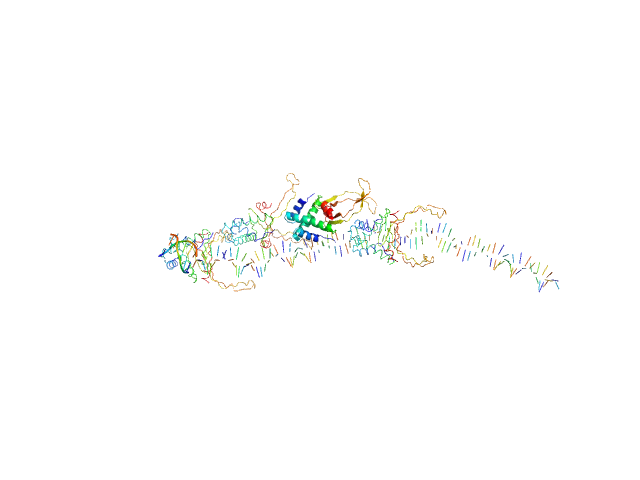
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E38K/V42L double mutant octamer, 76 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Apr 23
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling (supplementary)
Soumya G Remesh
|
| RgGuinier |
5.8 |
nm |
| Dmax |
28.1 |
nm |
| VolumePorod |
296 |
nm3 |
|
|
|
|
|
|
|
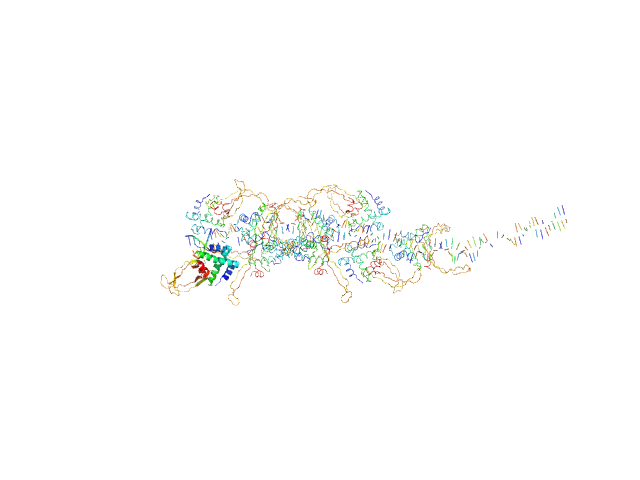
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E38K/V42L double mutant 16-mer, 153 kDa Linked to wild-type … protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Apr 23
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling (supplementary)
Soumya G Remesh
|
| RgGuinier |
6.3 |
nm |
| Dmax |
27.3 |
nm |
| VolumePorod |
401 |
nm3 |
|
|