|
|
|
|

|
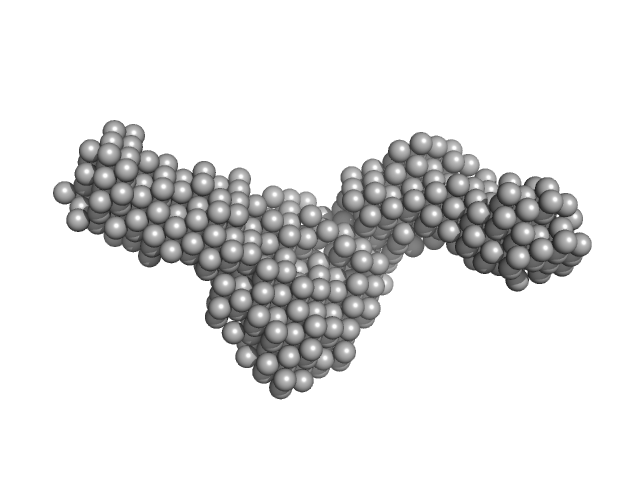
| Sample: |
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Nicked DNA dimer, 16 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 13
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Probable exodeoxyribonuclease III protein XthA monomer, 33 kDa Mycobacterium tuberculosis protein
Nicked DNA monomer, 8 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 13
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2015 May 24
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2015 Sep 21
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2015 Nov 23
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2019 Jul 21
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2015 May 23
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.6 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Apr 8
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Apr 8
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
231 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2015 May 24
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.6 |
nm |
| VolumePorod |
65 |
nm3 |
|
|