|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Humanized immunoglobulin G1 monoclonal antibody monomer, 148 kDa Mouse/human (chimera)
|
| Buffer: |
100 mM glycine-HCl, pH: 2 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2016 Mar 2
|
Getting Smaller by Denaturation: Acid-Induced Compaction of Antibodies
The Journal of Physical Chemistry Letters :3898-3906 (2023)
Imamura H, Ooishi A, Honda S
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Humanized immunoglobulin G1 monoclonal antibody monomer, 148 kDa Mouse/human (chimera)
|
| Buffer: |
100 mM glycine-HCl, 200 mM NaCl, pH: 2 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Mar 5
|
Getting Smaller by Denaturation: Acid-Induced Compaction of Antibodies
The Journal of Physical Chemistry Letters :3898-3906 (2023)
Imamura H, Ooishi A, Honda S
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
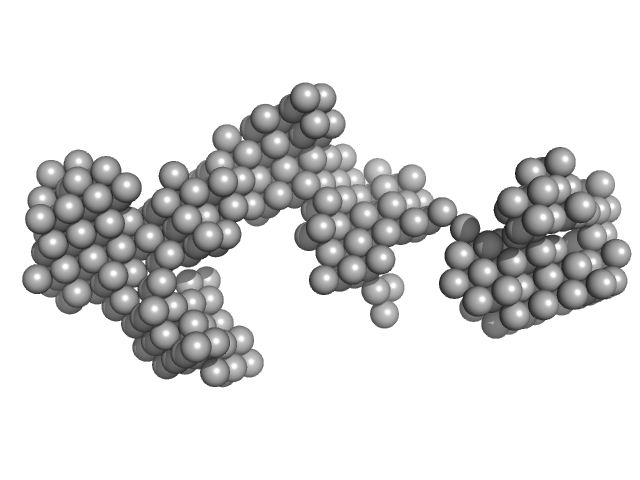
| Sample: |
Japanese encephaltitis 5' TR monomer, 74 kDa Japanese encephalitis virus RNA
|
| Buffer: |
10 mM Bis-tris, 100 mM NaCl, 15 mM KCl 15 mM MgCl2, 10% glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 22
|
Investigating RNA-RNA interactions through computational and biophysical analysis.
Nucleic Acids Res (2023)
Mrozowich T, Park SM, Waldl M, Henrickson A, Tersteeg S, Nelson CR, De Klerk A, Demeler B, Hofacker IL, Wolfinger MT, Patel TR
|
| RgGuinier |
7.0 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
356 |
nm3 |
|
|
|
|
|
|
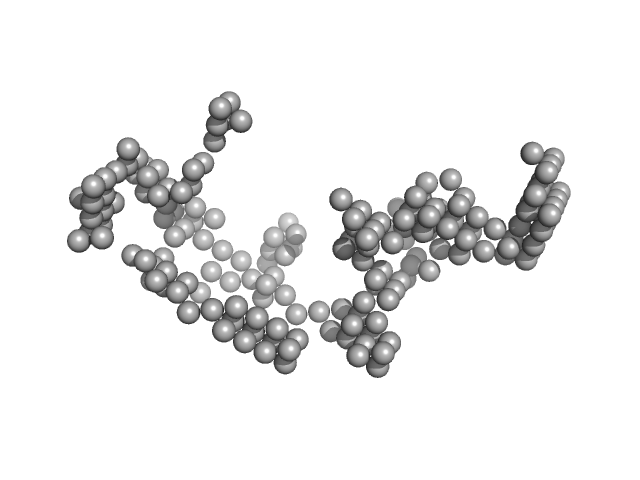
|
| Sample: |
Japanese encephalitis virus 3' UTR monomer, 186 kDa Japanese encephalitis virus RNA
|
| Buffer: |
10 mM Bis-tris, 100 mM NaCl, 15 mM KCl 15 mM MgCl2, 10% glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 22
|
Investigating RNA-RNA interactions through computational and biophysical analysis.
Nucleic Acids Res (2023)
Mrozowich T, Park SM, Waldl M, Henrickson A, Tersteeg S, Nelson CR, De Klerk A, Demeler B, Hofacker IL, Wolfinger MT, Patel TR
|
| RgGuinier |
11.3 |
nm |
| Dmax |
34.6 |
nm |
| VolumePorod |
2900 |
nm3 |
|
|