|
|
|
|
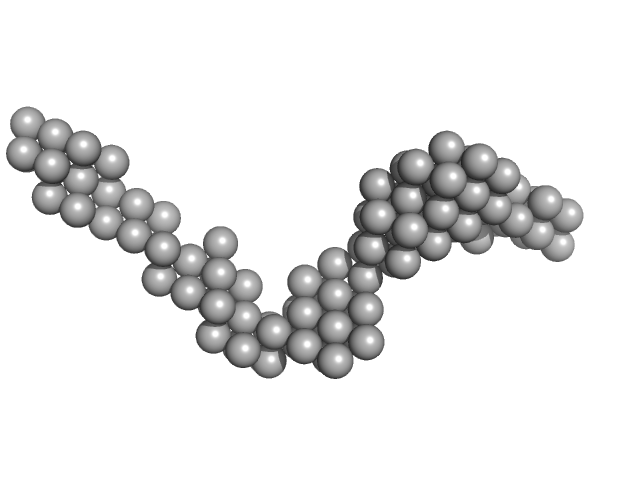
|
| Sample: |
Octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 4
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
7.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
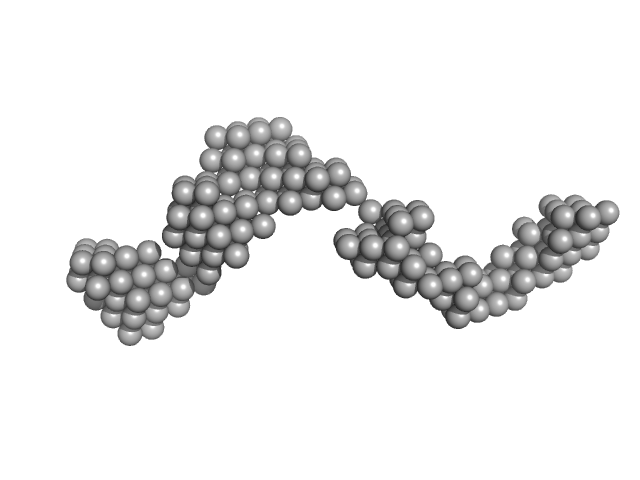
|
| Sample: |
Octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
8.8 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
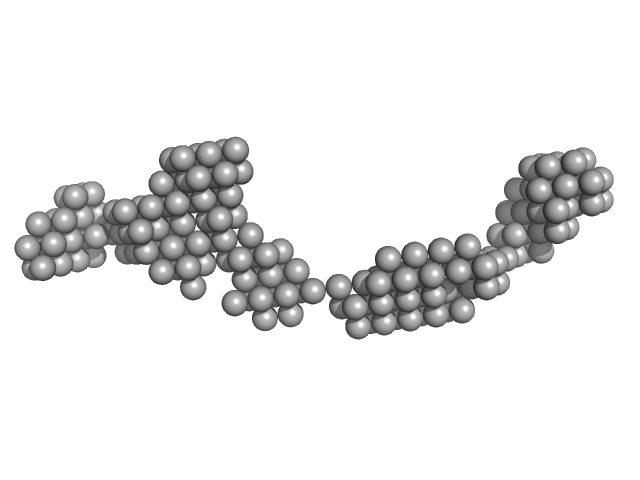
|
| Sample: |
Octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.0 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl and 1 mM PDS, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.1 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
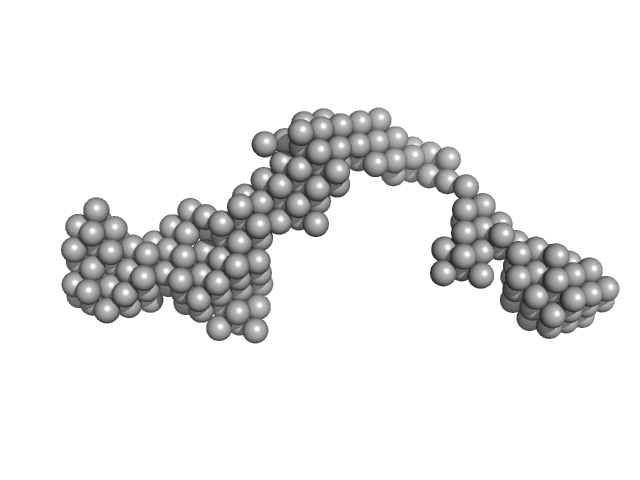
|
| Sample: |
Octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
7.0 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.0 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
8.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
223 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl and 1 mM PDS, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.2 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-L-Glutamic Acid, 6 kDa
|
| Buffer: |
Dimethyl Sulfoxide (DMSO), pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 11
|
β2-Type Amyloidlike Fibrils of Poly-l-glutamic Acid Convert into Long, Highly Ordered Helices upon Dissolution in Dimethyl Sulfoxide.
J Phys Chem B 122(50):11895-11905 (2018)
Berbeć S, Dec R, Molodenskiy D, Wielgus-Kutrowska B, Johannessen C, Hernik-Magoń A, Tobias F, Bzowska A, Ścibisz G, Keiderling TA, Svergun D, Dzwolak W
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Truncated P5abc subdomain from tetrahymena ribozyme monomer, 18 kDa RNA
|
| Buffer: |
10mM KMOPS 20mM KCl 1mM MgCl2 20uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 3
|
Revealing the distinct folding phases of an RNA three-helix junction.
Nucleic Acids Res 46(14):7354-7365 (2018)
Plumridge A, Katz AM, Calvey GD, Elber R, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
|
|