|
|
|
|
|
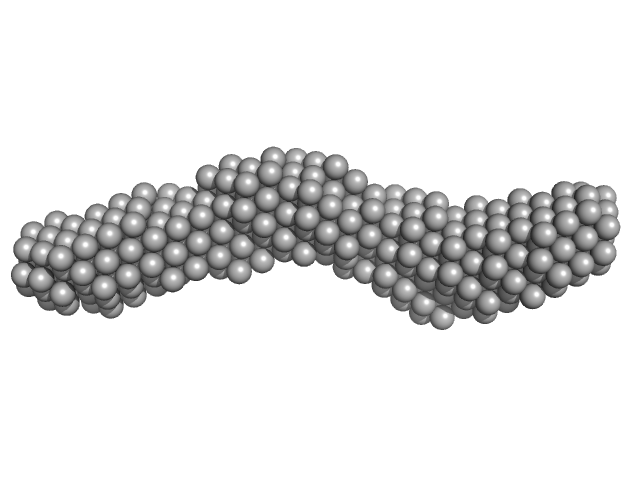
| Sample: |
3'SL from Zika virus monomer, 32 kDa Zika virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
3'SL from West Nile virus monomer, 31 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2017 Apr 7
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Subgenomic flavivirus RNA from Zika virus monomer, 133 kDa Zika virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Mar 13
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
7.5 |
nm |
| Dmax |
29.6 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Subgenomic flavivirus RNAs from Dengue virus 2 monomer, 137 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 16
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
7.9 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Subgenomic flavivirus RNAs from West nile virus monomer, 170 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Mar 13
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
7.9 |
nm |
| Dmax |
31.8 |
nm |
| VolumePorod |
261 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
J-DNA (23mer) monomer, 14 kDa DNA
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 21
|
The domain architecture of protozoan protein J-DNA-binding protein 1 suggests synergy between base J DNA binding and thymidine hydroxylase activity.
J Biol Chem (2019)
Adamopoulos A, Heidebrecht T, Roosendaal J, Touw WG, Phan IQ, Beijnen J, Perrakis A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RoX2 stem-loop 7, 18-mer fragment monomer, 12 kDa synthetic construct RNA
|
| Buffer: |
20 mM NaPO4, 200 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 29
|
Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless.
Nucleic Acids Res 47(8):4319-4333 (2019)
Ankush Jagtap PK, Müller M, Masiewicz P, von Bülow S, Hollmann NM, Chen PC, Simon B, Thomae AW, Becker PB, Hennig J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Metaphase chromosomes in 17 mM magnesium chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, 17 mM magnesium chloride, 20 mM sodium chloride, 120 mM potassium chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 May 23
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|
|
|
|
|
|
| Sample: |
Human metaphase chromosomes in 5 mM magnesium chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, 5 mM magnesium chloride, 10 mM sodium chloride, 120 mM potassium chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 Jun 6
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|
|
|
|
|
|
| Sample: |
Human metaphase chromosomes in 1 mM hexaamminecobalt(III) chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, hexaamminecobalt(III) chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 Sep 12
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|