|
|
|
|

|
| Sample: |
At3g55760 monomer, 60 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Dec 8
|
LIKE EARLY STARVATION 1 and EARLY STARVATION 1 promote and stabilize amylopectin phase transition in starch biosynthesis.
Sci Adv 9(21):eadg7448 (2023)
Liu C, Pfister B, Osman R, Ritter M, Heutinck A, Sharma M, Eicke S, Fischer-Stettler M, Seung D, Bompard C, Abt MR, Zeeman SC
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Inactive purple acid phosphatase-like protein monomer, 40 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Dec 8
|
LIKE EARLY STARVATION 1 and EARLY STARVATION 1 promote and stabilize amylopectin phase transition in starch biosynthesis.
Sci Adv 9(21):eadg7448 (2023)
Liu C, Pfister B, Osman R, Ritter M, Heutinck A, Sharma M, Eicke S, Fischer-Stettler M, Seung D, Bompard C, Abt MR, Zeeman SC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CP12-2 (D77G) monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
25 mM potassium phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Conformational Disorder Analysis of the Conditionally Disordered Protein CP12 from Arabidopsis thaliana in Its Different Redox States
International Journal of Molecular Sciences 24(11):9308 (2023)
Del Giudice A, Gurrieri L, Galantini L, Fanti S, Trost P, Sparla F, Fermani S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CP12-2 (D77G) monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
25 mM potassium phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Conformational Disorder Analysis of the Conditionally Disordered Protein CP12 from Arabidopsis thaliana in Its Different Redox States
International Journal of Molecular Sciences 24(11):9308 (2023)
Del Giudice A, Gurrieri L, Galantini L, Fanti S, Trost P, Sparla F, Fermani S
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
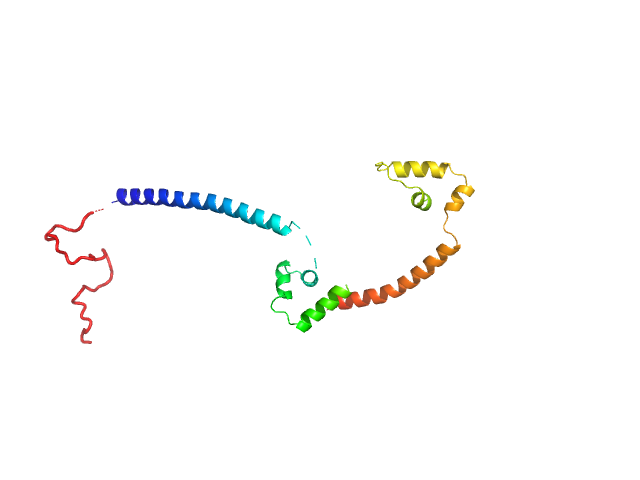
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
|
| Buffer: |
50mM Tris, 750mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Oct 10
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 9
|
Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial β-oxidation trifunctional enzymes.
Structure (2023)
Sah-Teli SK, Pinkas M, Hynönen MJ, Butcher SJ, Wierenga RK, Novacek J, Venkatesan R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial monomer, 14 kDa Homo sapiens protein
NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial monomer, 14 kDa Homo sapiens protein
Iron-sulfur cluster assembly 2 homolog, mitochondrial monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
43 |
nm3 |
|
|