|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase monomer, 150 kDa Saccharomyces cerevisiae protein
Mitochondrial transcription factor 1 monomer, 40 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM DDT, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Dec 7
|
PfKsgA1 functions as a transcription initiation factor and interacts with the N-terminal region of the mitochondrial RNA polymerase of Plasmodium falciparum.
Int J Parasitol (2020)
Gupta A, Shrivastava D, Shakya AK, Gupta K, Pratap JV, Habib S
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
292 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aldehyde-alcohol dehydrogenase dimer, 97 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris 400 mM NaCl 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Nov 27
|
High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):414-421 (2020)
Azmi L, Bragginton EC, Cadby IT, Byron O, Roe AJ, Lovering AL, Gabrielsen M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aldehyde-alcohol dehydrogenase monomer, 48 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
30 mM HEPES, 150 mM NaCl, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 30
|
High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):414-421 (2020)
Azmi L, Bragginton EC, Cadby IT, Byron O, Roe AJ, Lovering AL, Gabrielsen M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleotide pyrophosphatase/phosphodiesterase from E. characias latex partially sequenced dimer, 28 kDa Euphorbia characias protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Feb 20
|
Structure of a nucleotide pyrophosphatase/phosphodiesterase (NPP) from Euphorbia characias latex characterized by small-angle X-ray scattering: clues for the general organization of plant NPPs.
Acta Crystallogr D Struct Biol 76(Pt 9):857-867 (2020)
Sabatucci A, Pintus F, Cabras T, Vincenzoni F, Maccarrone M, Medda R, Dainese E
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
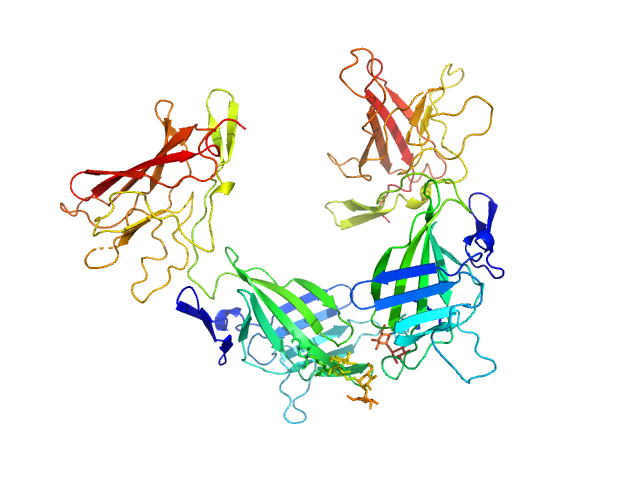
|
| Sample: |
Cation-independent mannose-6-phosphate receptor dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
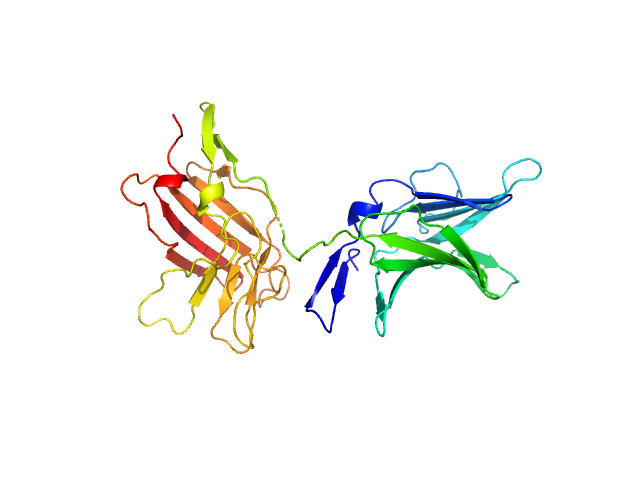
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
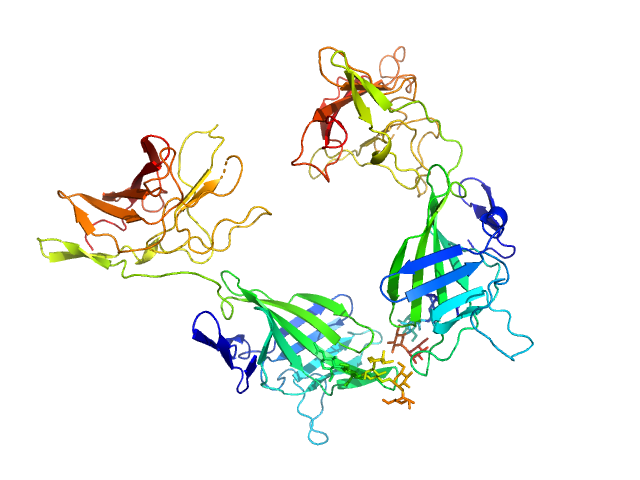
|
| Sample: |
Cation-independent mannose-6-phosphate receptor dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
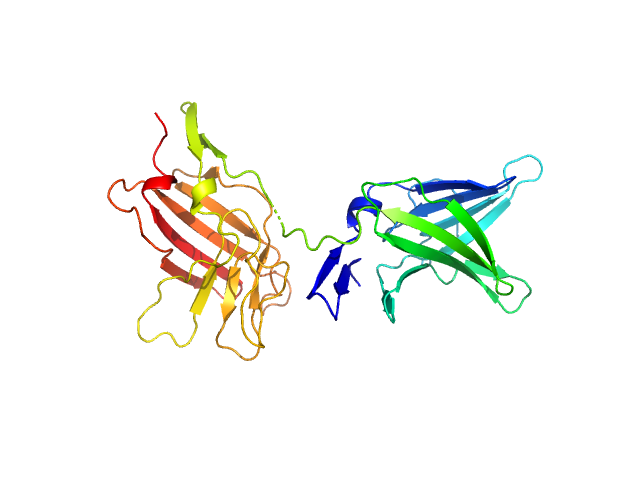
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
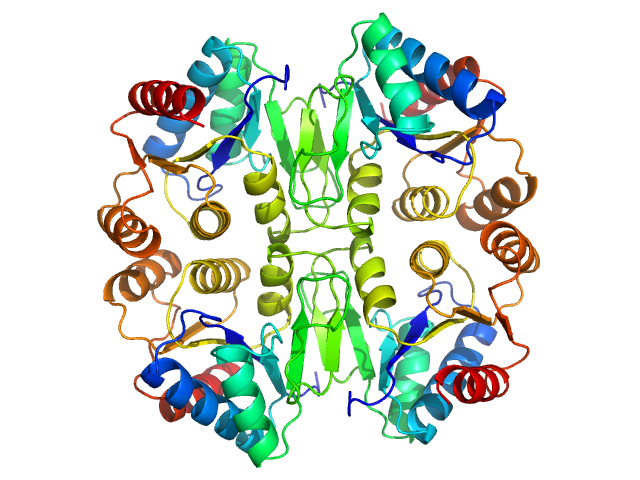
| Sample: |
P450 cytochrome, putative (Moco carrier protein) tetramer, 75 kDa Rippkaea orientalis (strain … protein
|
| Buffer: |
100 mM Tris-HCl, 300 mM NaCl, 2 %(v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 5
|
The structure of the Moco carrier protein from Rippkaea orientalis.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):453-463 (2020)
Krausze J, Hercher TW, Archna A, Kruse T
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type 3 secretion system pilotin dimer, 28 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Mar 23
|
Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure (2020)
Majewski DD, Okon M, Heinkel F, Robb CS, Vuckovic M, McIntosh LP, Strynadka NCJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
53 |
nm3 |
|
|