|
|
|
|

|
| Sample: |
Type 3 secretion system pilotin dimer, 19 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Mar 23
|
Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure (2020)
Majewski DD, Okon M, Heinkel F, Robb CS, Vuckovic M, McIntosh LP, Strynadka NCJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Vitamin K-dependent protein C monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 145 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 14
|
Zymogen and activated protein C have similar structural architecture.
J Biol Chem (2020)
Stojanovski BM, Pelc LA, Zuo X, Di Cera E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|
|
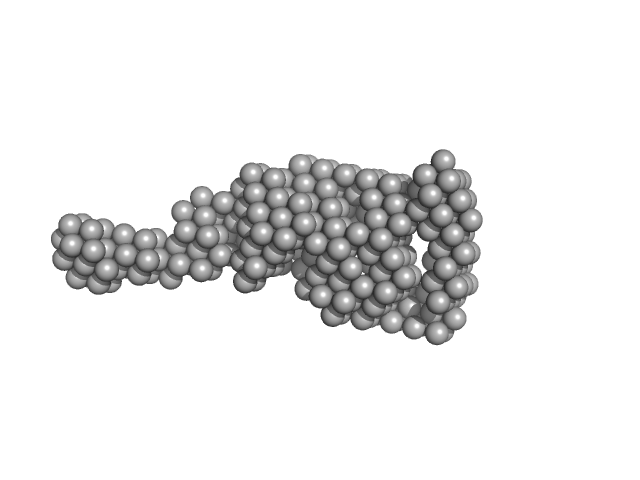
| Sample: |
Vitamin K-dependent protein C monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 145 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Aug 17
|
Zymogen and activated protein C have similar structural architecture.
J Biol Chem (2020)
Stojanovski BM, Pelc LA, Zuo X, Di Cera E
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 57 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 45 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 32 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
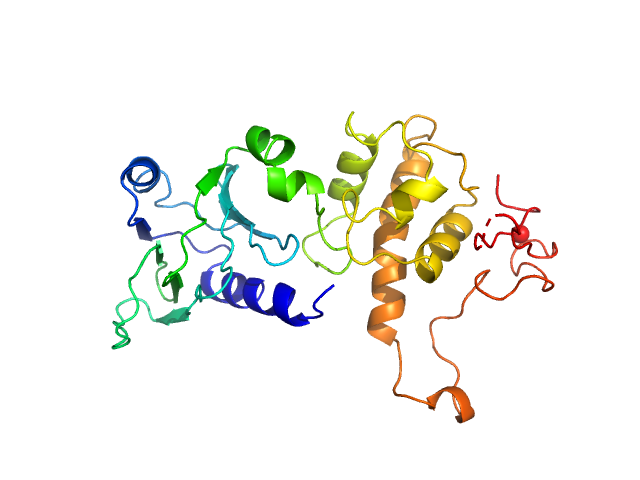
| Sample: |
Endonuclease 8 monomer, 31 kDa Escherichia coli protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
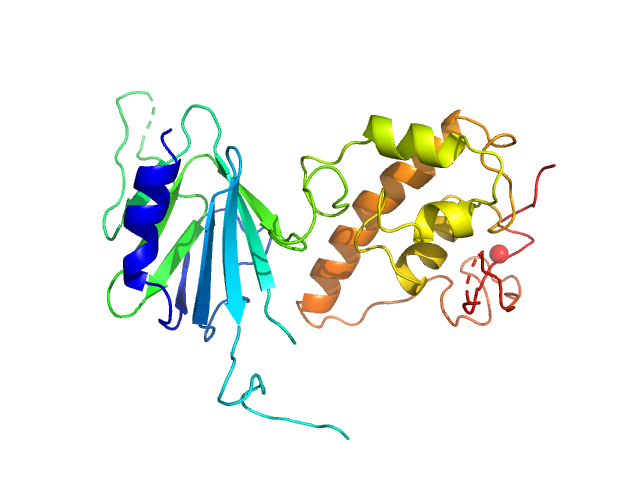
| Sample: |
Nei like DNA glycosylase 2 monomer, 39 kDa Monodelphis domestica protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
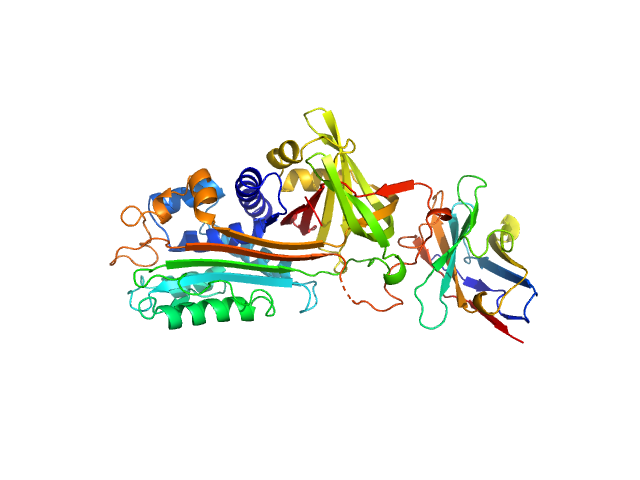
| Sample: |
Nei like DNA glycosylase 2 (Δ67-133) monomer, 33 kDa Monodelphis domestica protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Plasminogen activator inhibitor 1 monomer, 43 kDa Homo sapiens protein
VHH-s-a93 (Ig module Nb93) monomer, 13 kDa Vicugna pacos protein
|
| Buffer: |
30 mM BIS-TRIS pH 5.5, 300 mM sodium chloride, 5% v/v glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 5
|
Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity
International Journal of Molecular Sciences 21(16):5859 (2020)
Sillen M, Weeks S, Strelkov S, Declerck P
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
80 |
nm3 |
|
|