|
|
|
|

|
| Sample: |
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP), mutation L664C monomer, 10 kDa Mus musculus protein
1-{4-[4-chloro-3-(trifluoromethyl)phenyl]-4-hydroxypiperidin-1-yl}-3-sulfanylpropan-1-one monomer, 0 kDa
|
| Buffer: |
25 mM Hepes, 150 NaCl, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
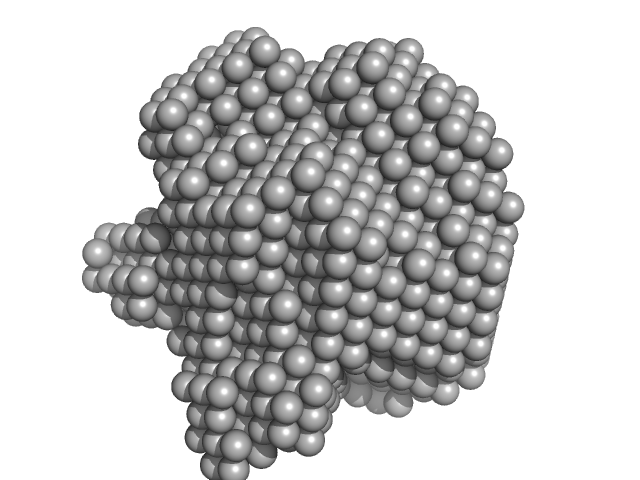
Bifunctional protein PaaZ hexamer, 438 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Feb 24
|
Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun 10(1):4127 (2019)
Sathyanarayanan N, Cannone G, Gakhar L, Katagihallimath N, Sowdhamini R, Ramaswamy S, Vinothkumar KR
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
636 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO SpaO(SPOA2) dimer, 25 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO SpaO(SPOA2) dimer, 25 kDa Salmonella enterica subsp. … protein
Surface presentation of antigens protein SpaO(SPOA1,2) monomer, 34 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO(SPOA1,2) N-terminus monomer, 17 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO SpaO(SPOA2) dimer, 25 kDa Salmonella enterica subsp. … protein
Surface presentation of antigens protein SpaO(SPOA1,2) N-terminus monomer, 17 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO SpaO(SPOA2) dimer, 25 kDa Salmonella enterica subsp. … protein
Surface presentation of antigens protein SpaO(SPOA1,2) monomer, 34 kDa Salmonella enterica subsp. … protein
Oxygen-regulated invasion protein OrgB dimer, 53 kDa Salmonella enterica subsp. … protein
ATP synthase InvC monomer, 48 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
10 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 12
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
6.0 |
nm |
| Dmax |
22.7 |
nm |
| VolumePorod |
302 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO(SPOA1,2) C-terminus monomer, 19 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
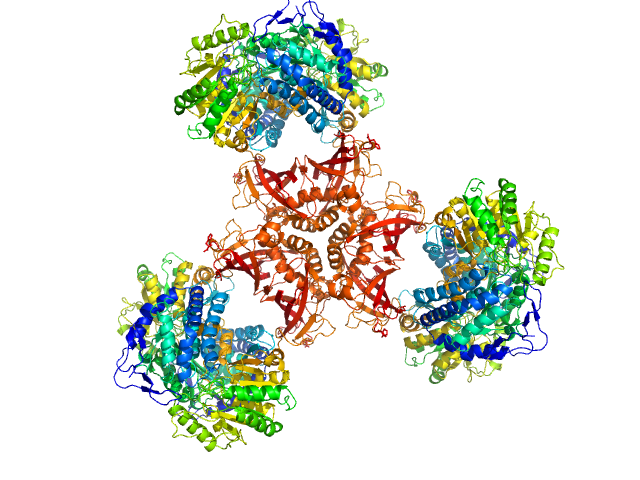
Methylxanthine N1-demethylase NdmA trimer, 127 kDa Pseudomonas putida protein
Methylxanthine N3-demethylase NdmB trimer, 129 kDa Pseudomonas putida protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 2 mM TCEP 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2018 Jul 27
|
Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J Mol Biol 431(19):3647-3661 (2019)
Kim JH, Kim BH, Brooks S, Kang SY, Summers RM, Song HK
|
| RgGuinier |
4.5 |
nm |
| Dmax |
12.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
Methylxanthine N1-demethylase NdmA trimer, 207 kDa Pseudomonas putida protein
Methylxanthine N3-demethylase NdmB trimer, 129 kDa Pseudomonas putida protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 2 mM TCEP 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2018 Jul 27
|
Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J Mol Biol 431(19):3647-3661 (2019)
Kim JH, Kim BH, Brooks S, Kang SY, Summers RM, Song HK
|
| RgGuinier |
5.4 |
nm |
| Dmax |
13.8 |
nm |
|
|