|
|
|
|
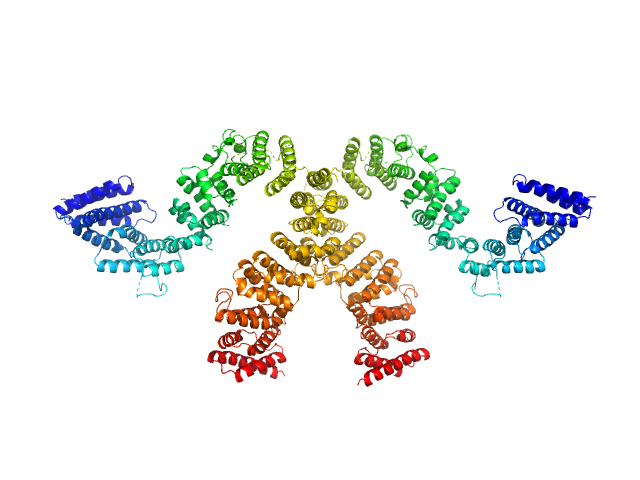
|
| Sample: |
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
463 |
nm3 |
|
|
|
|
|
|
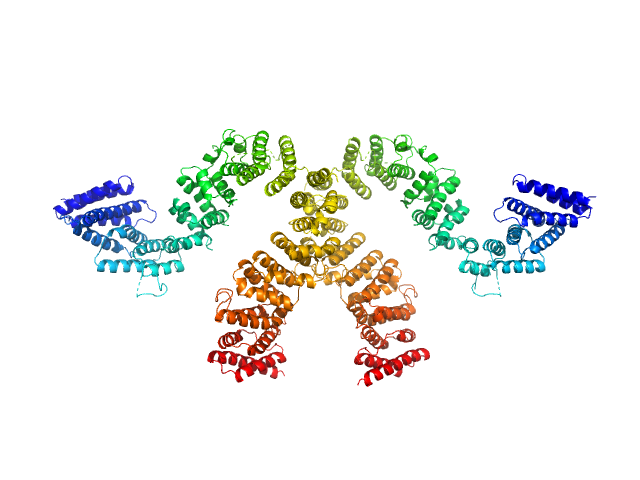
|
| Sample: |
Condensin complex subunit 3-like protein monomer, 108 kDa Chaetomium thermophilum protein
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
5.3 |
nm |
| Dmax |
18.7 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
|
|
|
|
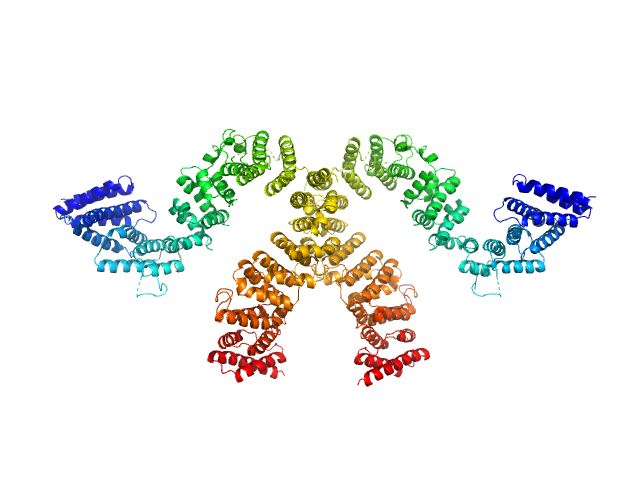
|
| Sample: |
Condensin complex subunit 3-like protein monomer, 108 kDa Chaetomium thermophilum protein
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
301 |
nm3 |
|
|
|
|
|
|
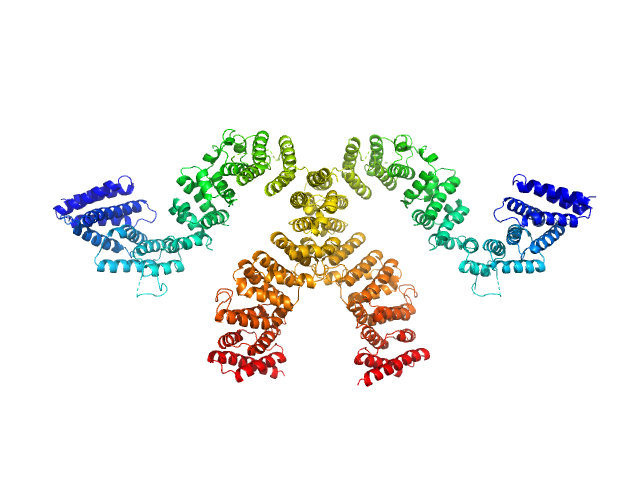
|
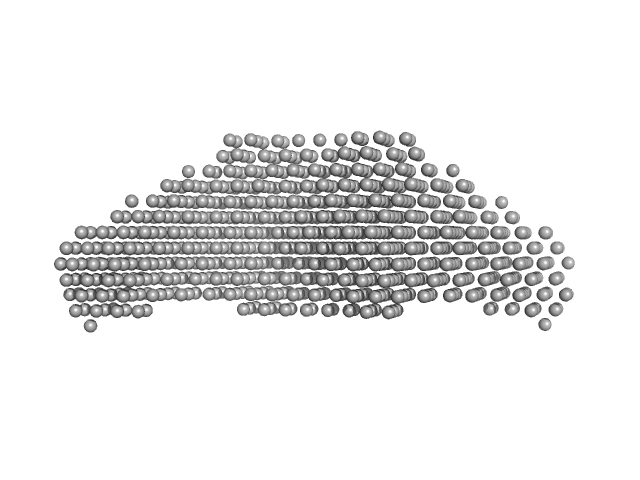
| Sample: |
Condensin complex subunit 3-like protein monomer, 108 kDa Chaetomium thermophilum protein
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
STI1-like protein dimer, 136 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2016 Aug 3
|
Structural studies of the Hsp70/Hsp90 organizing protein of Plasmodium falciparum and its modulation of Hsp70 and Hsp90 ATPase activities.
Biochim Biophys Acta Proteins Proteom :140282 (2019)
Silva NSM, Bertolino-Reis DE, Dores-Silva PR, Anneta FB, Seraphim TV, Barbosa LRS, Borges JC
|
| RgGuinier |
6.3 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
557 |
nm3 |
|
|
|
|
|
|
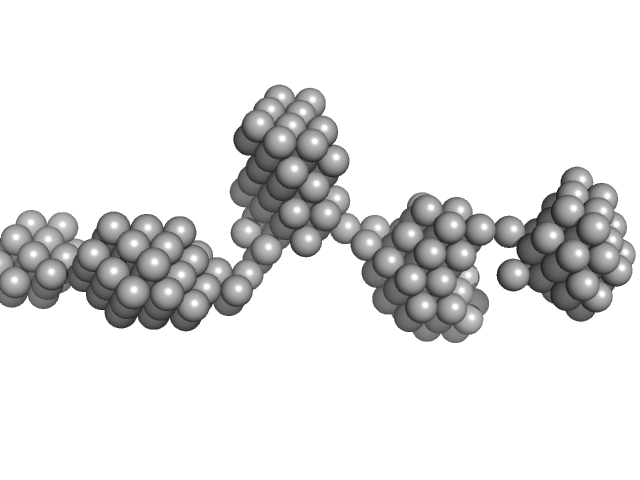
|
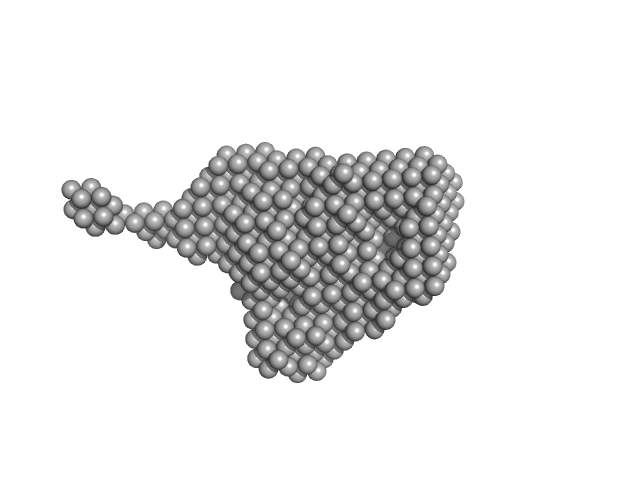
| Sample: |
Aryl hydrocarbon receptor nuclear translocator-like protein 1 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP), mutation L664C monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
1.9 |
nm |
| Dmax |
8.1 |
nm |
|
|
|
|
|
|

|
| Sample: |
Aryl hydrocarbon receptor nuclear translocator-like protein 1 monomer, 10 kDa Mus musculus protein
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP), mutation L664C monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Brain and muscle ARNT-like 1 (G517-L625) including transactivation domain (TAD) monomer, 11 kDa Mus musculus protein
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP) monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP) monomer, 10 kDa Mus musculus protein
Brain and muscle ARNT-like 1 (G490-L625) including transactivation domain (TAD) monomer, 14 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
17.5 |
nm |
|
|