|
|
|
|
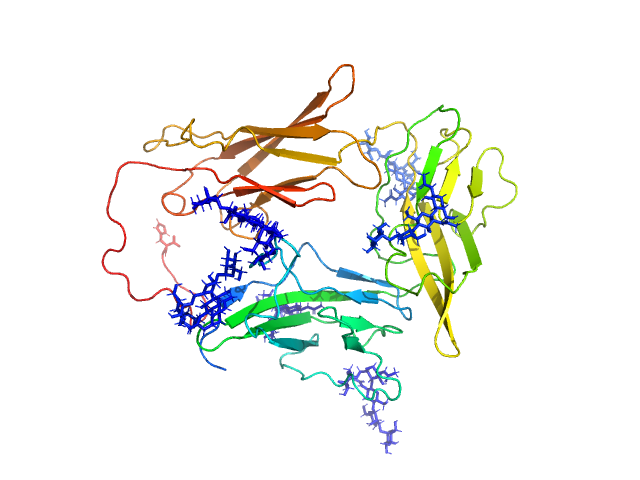
|
| Sample: |
Interleukin-1 receptor accessory protein ectodomain with RI linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Aug 11
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
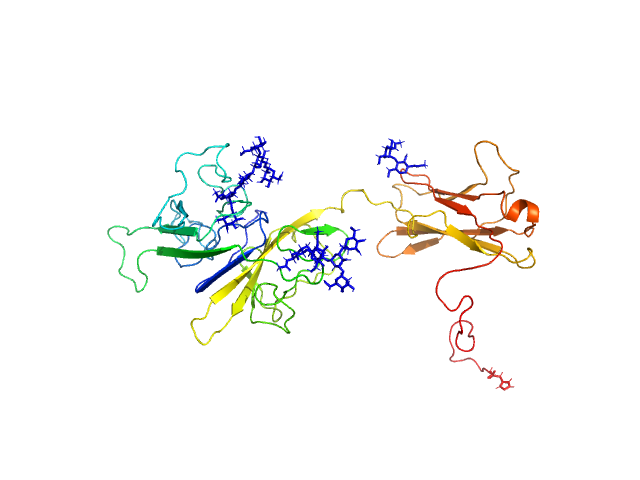
|
| Sample: |
Interleukin-18 receptor accessory protein ectodomain with Rα linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
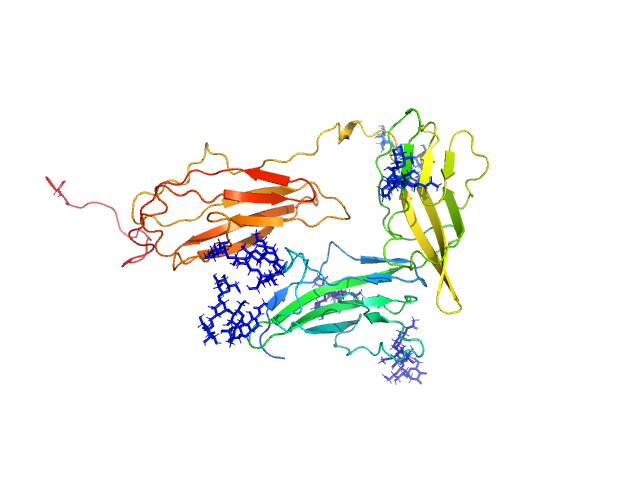
|
| Sample: |
Interleukin-1 receptor accessory protein ectodomains with ST2 linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
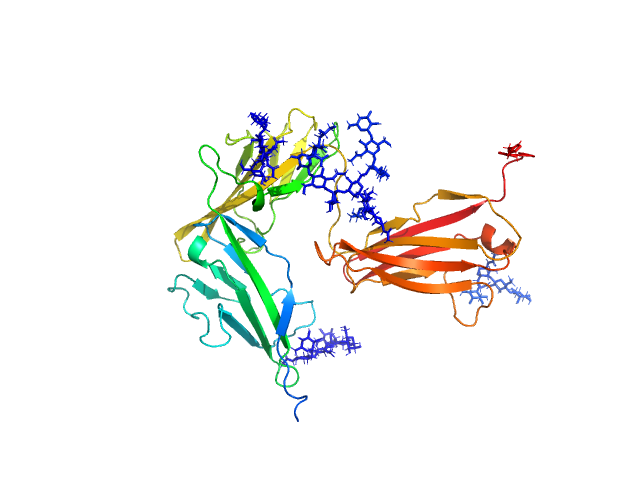
|
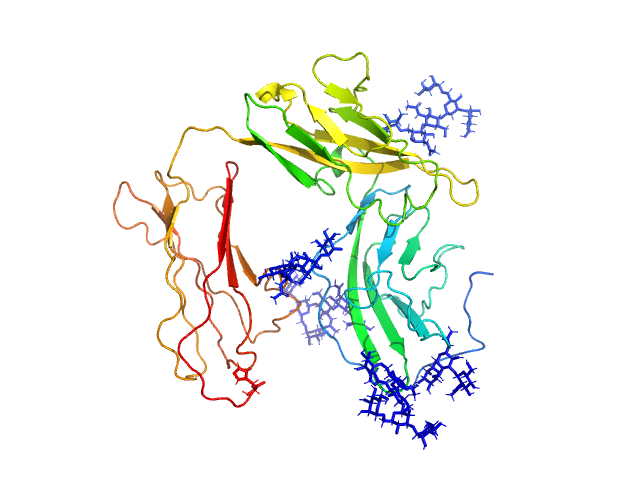
| Sample: |
Interleukin-1 receptor type 1 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 22
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
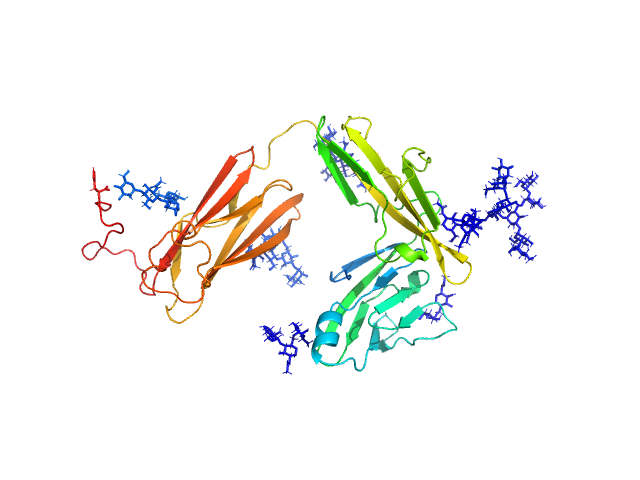
| Sample: |
Interleukin-1 receptor type 2 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
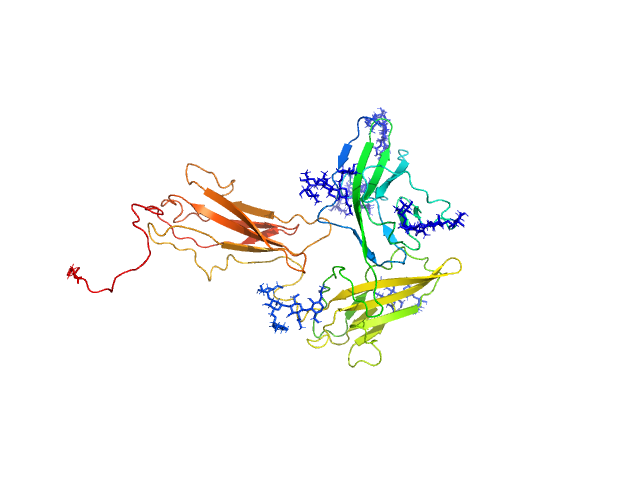
| Sample: |
Interleukin-18 receptor 1 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Oct 3
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
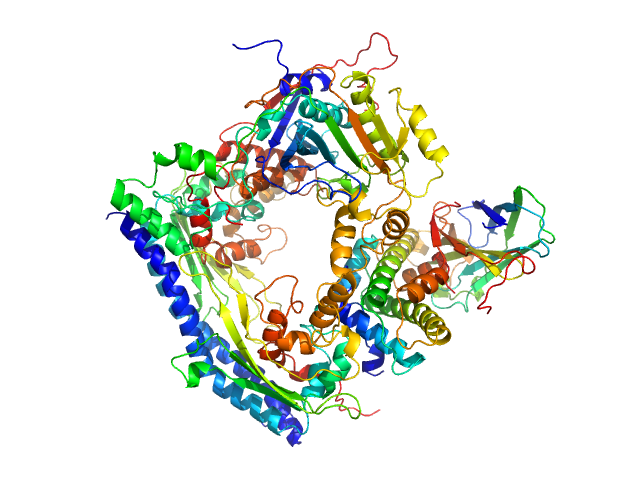
| Sample: |
Interleukin-1 receptor accessory protein ectodomains with RII linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 3
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.8 |
nm |
|
|
|
|
|
|
|
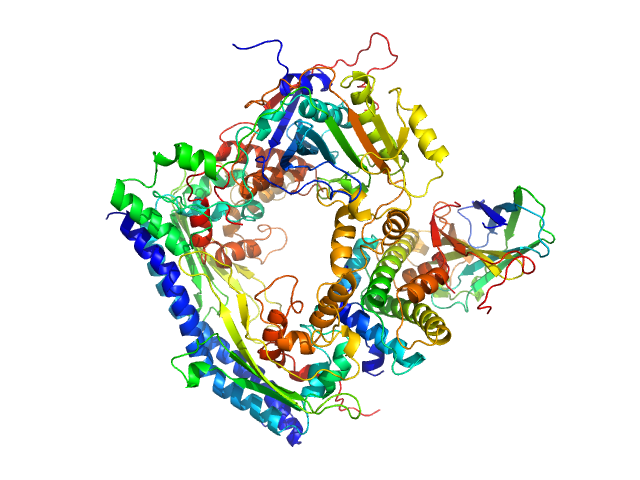
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 4
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
|
|
|
|
|
|
|
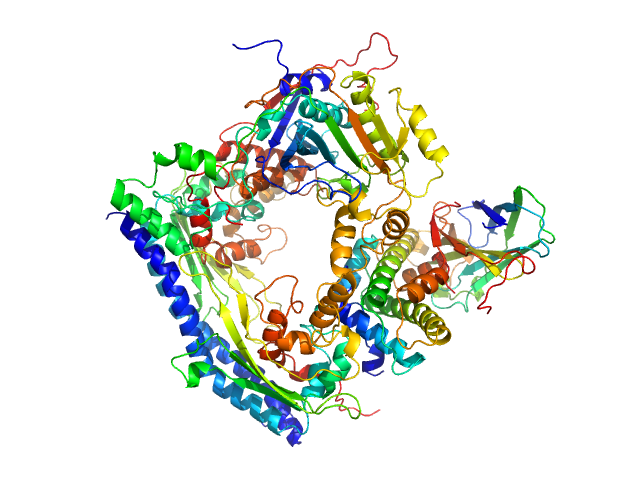
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 4
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
|
|