|
|
|
|
|
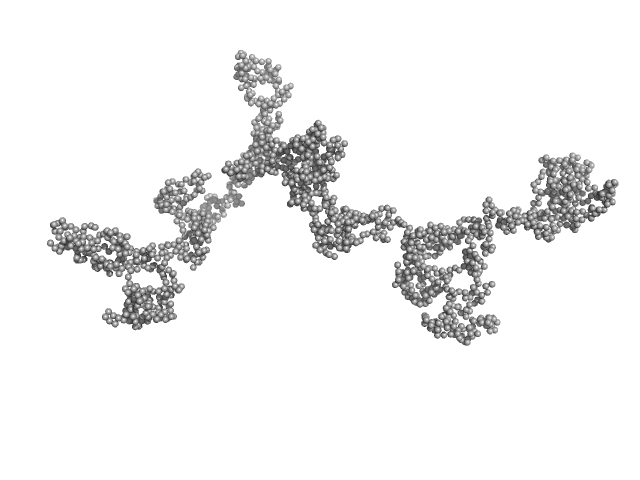
| Sample: |
Transcription intermediary factor 1-beta, TIF1b, KAP1, TRIM28 dimer, 183 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 6
|
KAP1 is an antiparallel dimer with a functional asymmetry.
Life Sci Alliance 2(4) (2019)
Fonti G, Marcaida MJ, Bryan LC, Träger S, Kalantzi AS, Helleboid PJ, Demurtas D, Tully MD, Grudinin S, Trono D, Fierz B, Dal Peraro M
|
| RgGuinier |
9.0 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
710 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Transcription intermediary factor 1-beta, TIF1b, KAP1, TRIM28, Fragment 23-418, RBCC domain dimer, 92 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 6
|
KAP1 is an antiparallel dimer with a functional asymmetry.
Life Sci Alliance 2(4) (2019)
Fonti G, Marcaida MJ, Bryan LC, Träger S, Kalantzi AS, Helleboid PJ, Demurtas D, Tully MD, Grudinin S, Trono D, Fierz B, Dal Peraro M
|
| RgGuinier |
8.3 |
nm |
| Dmax |
37.0 |
nm |
| VolumePorod |
381 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transcription intermediary factor 1-beta, TIF1b, KAP1, TRIM28; amino acids 23-812 dimer, 175 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 6
|
KAP1 is an antiparallel dimer with a functional asymmetry.
Life Sci Alliance 2(4) (2019)
Fonti G, Marcaida MJ, Bryan LC, Träger S, Kalantzi AS, Helleboid PJ, Demurtas D, Tully MD, Grudinin S, Trono D, Fierz B, Dal Peraro M
|
| RgGuinier |
8.9 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
853 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Filamin A Ig-like domains 4-6 monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 10
|
Critical Structural Defects Explain Filamin A Mutations Causing Mitral Valve Dysplasia.
Biophys J 117(8):1467-1475 (2019)
Haataja TJK, Capoulade R, Lecointe S, Hellman M, Merot J, Permi P, Pentikäinen U
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Filamin A Ig-like domains 4-6, V711D mutation monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 10
|
Critical Structural Defects Explain Filamin A Mutations Causing Mitral Valve Dysplasia.
Biophys J 117(8):1467-1475 (2019)
Haataja TJK, Capoulade R, Lecointe S, Hellman M, Merot J, Permi P, Pentikäinen U
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Filamin A Ig-like domains 4-6, V711D mutation monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 10
|
Critical Structural Defects Explain Filamin A Mutations Causing Mitral Valve Dysplasia.
Biophys J 117(8):1467-1475 (2019)
Haataja TJK, Capoulade R, Lecointe S, Hellman M, Merot J, Permi P, Pentikäinen U
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Filamin A Ig-like domains 4-6, H743P mutation monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 10
|
Critical Structural Defects Explain Filamin A Mutations Causing Mitral Valve Dysplasia.
Biophys J 117(8):1467-1475 (2019)
Haataja TJK, Capoulade R, Lecointe S, Hellman M, Merot J, Permi P, Pentikäinen U
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Filamin A Ig-like domains 4-6, H743P mutation monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 10
|
Critical Structural Defects Explain Filamin A Mutations Causing Mitral Valve Dysplasia.
Biophys J 117(8):1467-1475 (2019)
Haataja TJK, Capoulade R, Lecointe S, Hellman M, Merot J, Permi P, Pentikäinen U
|
| RgGuinier |
4.0 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histidine-binding periplasmic protein monomer, 26 kDa Escherichia coli protein
|
| Buffer: |
100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 10
|
Structure-based screening of binding affinities via small-angle X-ray scattering
(2019)
Chen P, Masiewicz P, Perez K, Hennig J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histidine-binding periplasmic protein monomer, 26 kDa Escherichia coli protein
|
| Buffer: |
100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 10
|
Structure-based screening of binding affinities via small-angle X-ray scattering
(2019)
Chen P, Masiewicz P, Perez K, Hennig J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
33 |
nm3 |
|
|