|
|
|
|
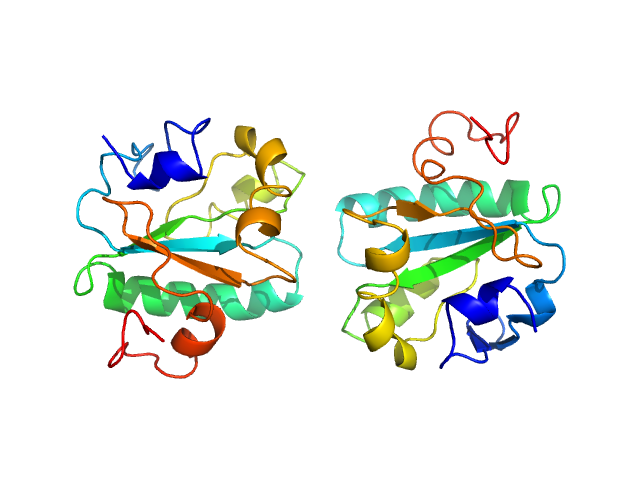
|
| Sample: |
Tryparedoxin K102E monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
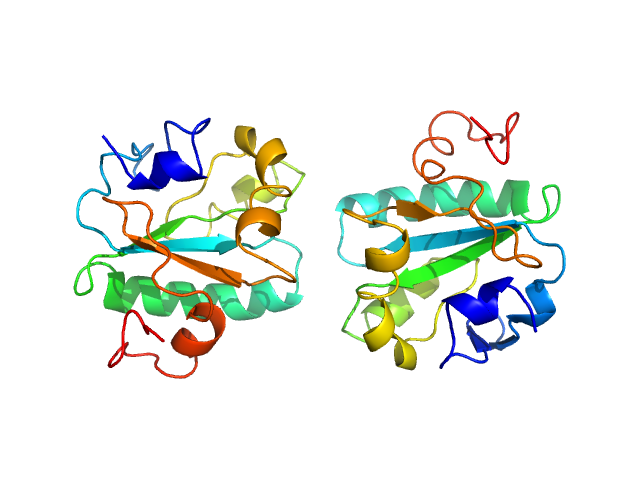
|
| Sample: |
Tryparedoxin K102E monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
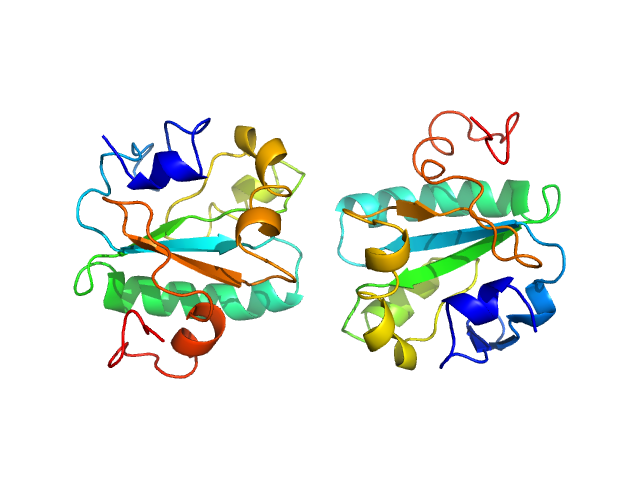
|
| Sample: |
Tryparedoxin K102E, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
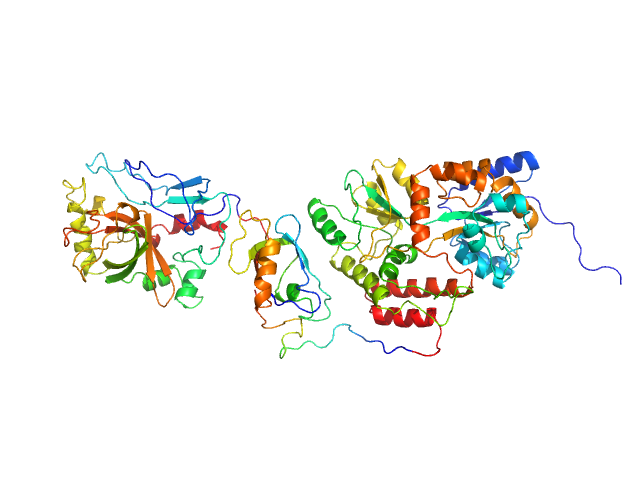
|
| Sample: |
SpoIVB peptidase (MBP fusion) monomer, 80 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Jul 12
|
Solution Structure of SpoIVB Reveals Mechanism of PDZ Domain-Regulated Protease Activity.
Front Microbiol 10:1232 (2019)
Xie X, Guo N, Xue G, Xie D, Yuan C, Harrison J, Li J, Jiang L, Huang M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein E2 tetramer, 139 kDa Homo sapiens protein
Heparin monomer, 15 kDa
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 3
|
Therapeutic approaches to ApoE
University of Sussex PhD thesis 2019 (2019)
Lucas Kraft
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
504 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein E2 tetramer, 139 kDa Homo sapiens protein
Heparin monomer, 15 kDa
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 3
|
Therapeutic approaches to ApoE
University of Sussex PhD thesis 2019 (2019)
Lucas Kraft
|
| RgGuinier |
6.4 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
531 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein E2 tetramer, 139 kDa Homo sapiens protein
Heparin monomer, 15 kDa
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 3
|
Therapeutic approaches to ApoE
University of Sussex PhD thesis 2019 (2019)
Lucas Kraft
|
| RgGuinier |
6.5 |
nm |
| Dmax |
23.4 |
nm |
| VolumePorod |
584 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein E2 tetramer, 139 kDa Homo sapiens protein
Heparin monomer, 15 kDa
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 3
|
Therapeutic approaches to ApoE
University of Sussex PhD thesis 2019 (2019)
Lucas Kraft
|
| RgGuinier |
7.1 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
666 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein E2 tetramer, 139 kDa Homo sapiens protein
Heparin monomer, 15 kDa
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 3
|
Therapeutic approaches to ApoE
University of Sussex PhD thesis 2019 (2019)
Lucas Kraft
|
| RgGuinier |
7.2 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
696 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDG25_fit1_model1.png)
|
| Sample: |
DNA repair protein RAD5 monomer, 138 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris, 150 mM KCl, 5 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Mar 6
|
Conformational flexibility of fork-remodeling helicase Rad5 shown by full-ensemble hybrid methods.
PLoS One 14(10):e0223875 (2019)
Gildenberg MS, Washington MT
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
254 |
nm3 |
|
|