|
|
|
|
|
| Sample: |
Small glutamine-rich tetratricopeptide repeat-containing protein alpha Nterminal-TPR domains dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 5
|
Structural complexity of the co-chaperone SGTA: a conserved C-terminal region is implicated in dimerization and substrate quality control.
BMC Biol 16(1):76 (2018)
Martínez-Lumbreras S, Krysztofinska EM, Thapaliya A, Spilotros A, Matak-Vinkovic D, Salvadori E, Roboti P, Nyathi Y, Muench JH, Roessler MM, Svergun DI, High S, Isaacson RL
|
|
|
|
|
|
|

|
| Sample: |
Designed Ankyrin Repeat Protein D1 monomer, 18 kDa synthetic construct protein
Kinesin-like protein KIF2A monomer, 48 kDa Homo sapiens protein
Tubulin alpha-1B chain dimer, 100 kDa Bos taurus protein
Tubulin beta-2B chain dimer, 100 kDa Bos taurus protein
|
| Buffer: |
HEPES 20 mM, MgCl2 1mM, NaCl 150mM, pH: 7.2 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 16
|
Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate.
Nat Commun 9(1):2628 (2018)
Trofimova D, Paydar M, Zara A, Talje L, Kwok BH, Allingham JS
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
374 |
nm3 |
|
|
|
|
|
|
|
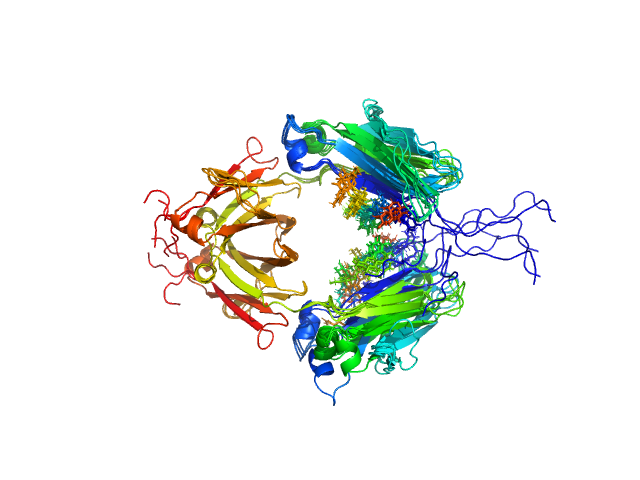
| Sample: |
Immunoglobulin heavy constant gamma 1 dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
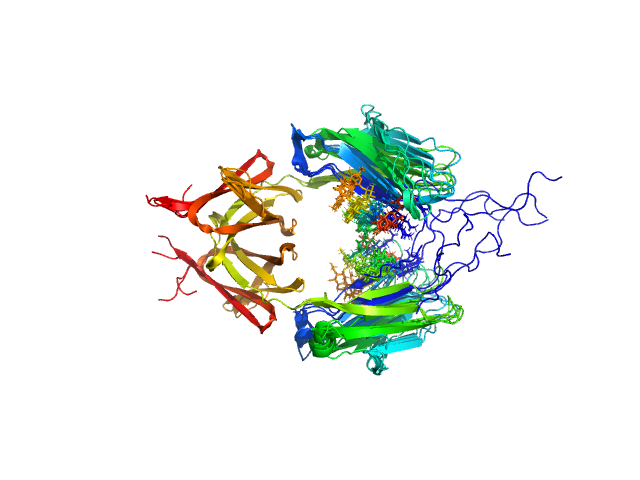
| Sample: |
Immunoglobulin heavy constant gamma 2 dimer, 52 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
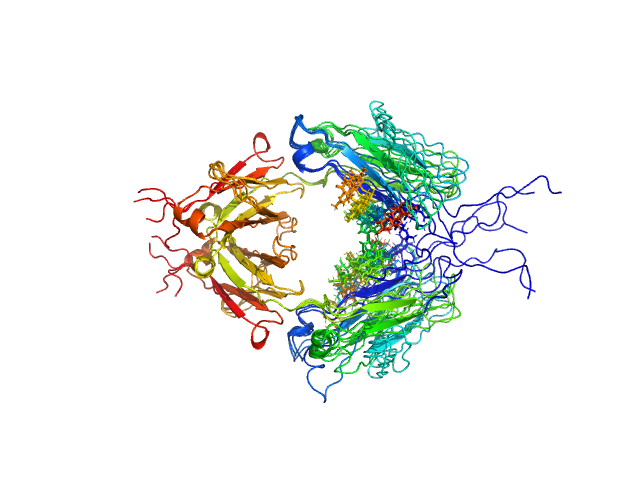
| Sample: |
Immunoglobulin heavy constant gamma 1 M255Y/S257T/T259E dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase from Clostridium botulinum tetramer, 126 kDa Clostridium botulinum protein
|
| Buffer: |
20mM Tris, 150mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2010 Nov 26
|
Substrate Locking Promotes Dimer-Dimer Docking of an Enzyme Antibiotic Target.
Structure 26(7):948-959.e5 (2018)
Atkinson SC, Dogovski C, Wood K, Griffin MDW, Gorman MA, Hor L, Reboul CF, Buckle AM, Wuttke J, Parker MW, Dobson RCJ, Perugini MA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase from Clostridium botulinum tetramer, 126 kDa Clostridium botulinum protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 5mM sodium pyruvate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2010 Nov 26
|
Substrate Locking Promotes Dimer-Dimer Docking of an Enzyme Antibiotic Target.
Structure 26(7):948-959.e5 (2018)
Atkinson SC, Dogovski C, Wood K, Griffin MDW, Gorman MA, Hor L, Reboul CF, Buckle AM, Wuttke J, Parker MW, Dobson RCJ, Perugini MA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
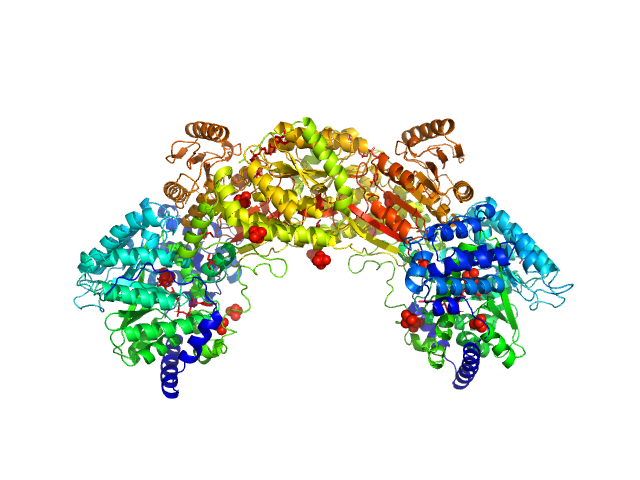
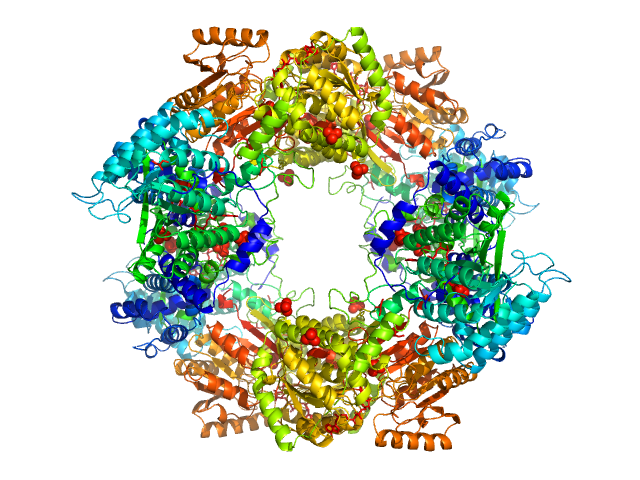
| Sample: |
Bifunctional protein PutA dimer, 215 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM TCEP, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 16
|
Redox Modulation of Oligomeric State in Proline Utilization A.
Biophys J 114(12):2833-2843 (2018)
Korasick DA, Campbell AC, Christgen SL, Chakravarthy S, White TA, Becker DF, Tanner JJ
|
| RgGuinier |
4.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
324 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bifunctional protein PutA tetramer, 430 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM TCEP, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 16
|
Redox Modulation of Oligomeric State in Proline Utilization A.
Biophys J 114(12):2833-2843 (2018)
Korasick DA, Campbell AC, Christgen SL, Chakravarthy S, White TA, Becker DF, Tanner JJ
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
582 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 4 dimer, 79 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 0.02% NaN3,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 13
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
183 |
nm3 |
|
|