|
|
|
|
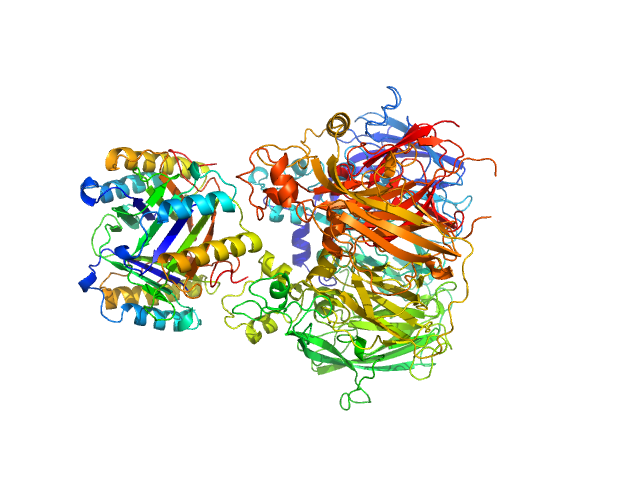
|
| Sample: |
Macrophage migration inhibitory factor trimer, 37 kDa Homo sapiens protein
Ceruloplasmin monomer, 122 kDa Homo sapiens protein
|
| Buffer: |
50mkm CuSO4, 100mM Hepes, pH: 7.4 |
| Experiment: |
SAXS
data collected at HECUS System-3, None on 2015 Dec 22
|
Structural Study of the Complex Formed by Ceruloplasmin and Macrophage Migration Inhibitory Factor.
Biochemistry (Mosc) 83(6):701-707 (2018)
Sokolov AV, Dadinova LA, Petoukhov MV, Bourenkov G, Dubova KM, Amarantov SV, Volkov VV, Kostevich VA, Gorbunov NP, Grudinina NA, Vasilyev VB, Samygina VR
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
228 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
CGMP-dependent protein kinase 1 monomer, 70 kDa Bos taurus protein
|
| Buffer: |
50 mM MES, 300 mM NaCl, 1 mM TCEP, 5 mM DTT, pH: 6.9 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Jun 6
|
An N-terminally truncated form of cyclic GMP-dependent protein kinase Iα (PKG Iα) is monomeric and autoinhibited and provides a model for activation.
J Biol Chem 293(21):7916-7929 (2018)
Moon TM, Sheehe JL, Nukareddy P, Nausch LW, Wohlfahrt J, Matthews DE, Blumenthal DK, Dostmann WR
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nonstructural protein sigma NS hexamer, 244 kDa Avian orthoreovirus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 25
|
Stability of local secondary structure determines selectivity of viral RNA chaperones.
Nucleic Acids Res (2018)
Bravo JPK, Borodavka A, Barth A, Calabrese AN, Mojzes P, Cockburn JJB, Lamb DC, Tuma R
|
| RgGuinier |
5.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
670 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Extender PN-Block (HL4) monomer, 27 kDa de novo protein protein
Stopper PN-Block (WA20) dimer, 25 kDa de novo protein protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 200 mM ArgHCl, 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2014 Dec 19
|
Self-Assembling Supramolecular Nanostructures Constructed from de Novo Extender Protein Nanobuilding Blocks.
ACS Synth Biol 7(5):1381-1394 (2018)
Kobayashi N, Inano K, Sasahara K, Sato T, Miyazawa K, Fukuma T, Hecht MH, Song C, Murata K, Arai R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|
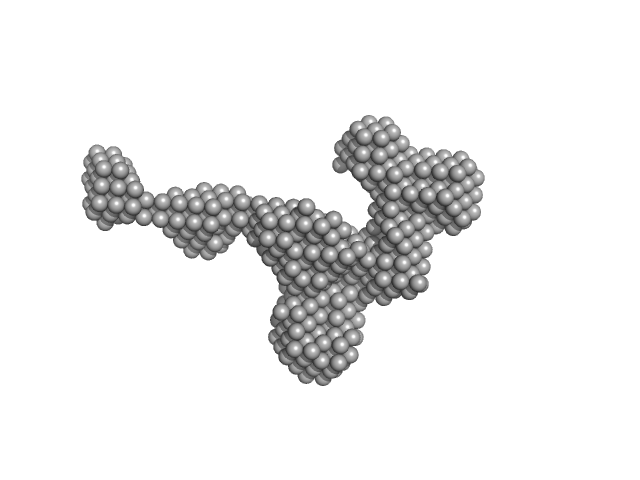
|
| Sample: |
Stopper PN-Block (WA20) dimer, 25 kDa de novo protein protein
Extender PN-Block (FL4) monomer, 27 kDa de novo protein protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 200 mM ArgHCl, 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2014 Dec 19
|
Self-Assembling Supramolecular Nanostructures Constructed from de Novo Extender Protein Nanobuilding Blocks.
ACS Synth Biol 7(5):1381-1394 (2018)
Kobayashi N, Inano K, Sasahara K, Sato T, Miyazawa K, Fukuma T, Hecht MH, Song C, Murata K, Arai R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
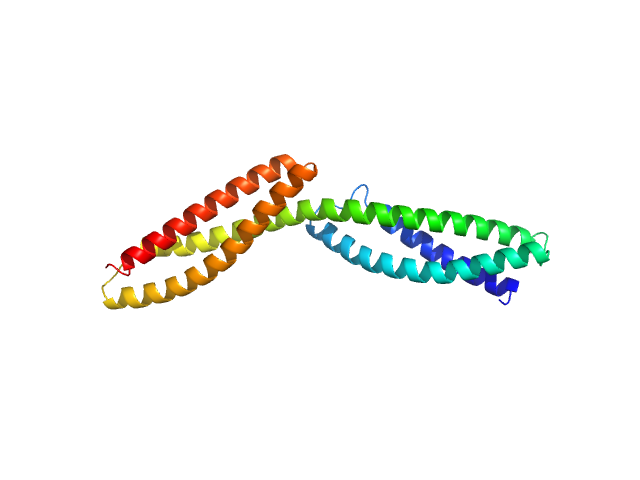
|
| Sample: |
Dystrophin central domain repeats 1 to 2 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Oct 7
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
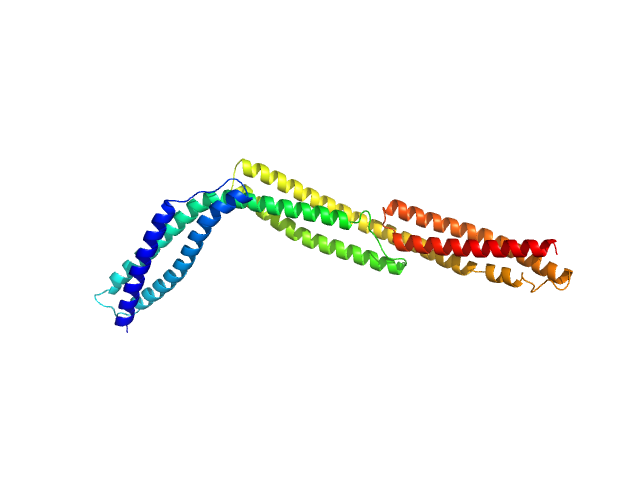
|
| Sample: |
Dystrophin central domain repeats 1 to 3 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Sep 9
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
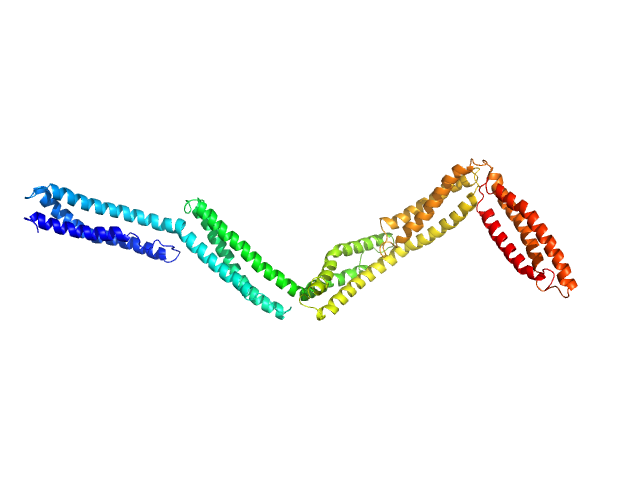
| Sample: |
Dystrophin central domain repeats 11 to 15. monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2011 Apr 11
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
5.8 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|

|
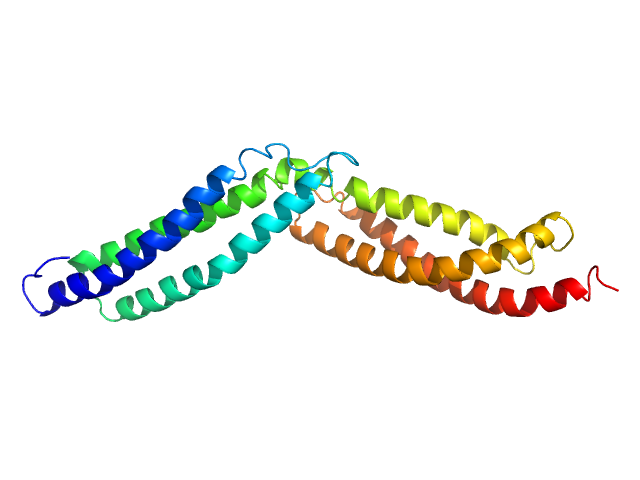
| Sample: |
Dystrophin central domain repeats 4 to 9 monomer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Sep 19
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dystrophin central domain repeats 16 to 17. monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Sep 19
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|