|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Lenalidomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa synthetic construct protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 29
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa synthetic construct protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 29
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 19
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
6.8 |
nm |
| Dmax |
30.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
High affinity immunoglobulin epsilon receptor subunit gamma monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
T-cell surface glycoprotein CD3 gamma chain monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
TYRO protein tyrosine kinase-binding protein monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
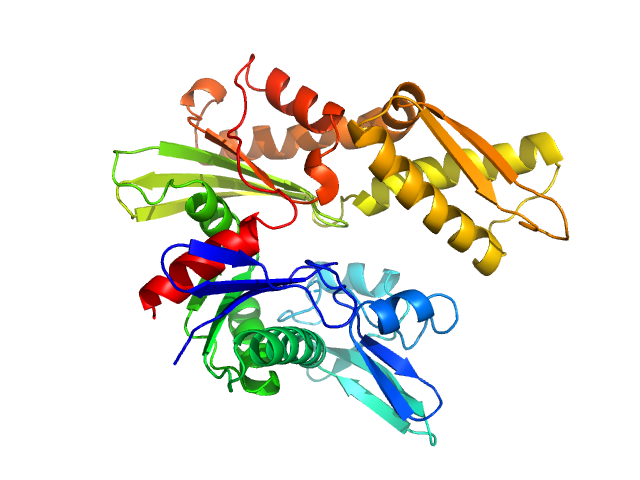
|
| Sample: |
Endoplasmic reticulum chaperone BiP monomer, 42 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 21
|
Structural basis of CDNF interaction with the UPR regulator GRP78.
Nat Commun 15(1):8175 (2024)
Graewert MA, Volkova M, Jonasson K, Määttä JAE, Gräwert T, Mamidi S, Kulesskaya N, Evenäs J, Johnsson RE, Svergun D, Bhattacharjee A, Huttunen HJ
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
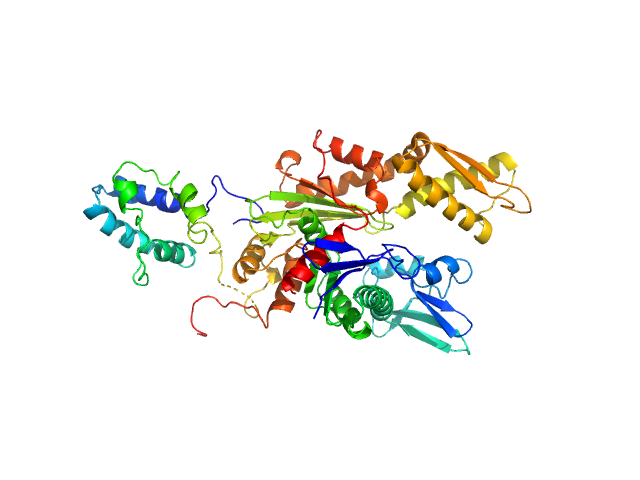
|
| Sample: |
Endoplasmic reticulum chaperone BiP monomer, 42 kDa Homo sapiens protein
Cerebral dopamine neurotrophic factor monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 21
|
Structural basis of CDNF interaction with the UPR regulator GRP78.
Nat Commun 15(1):8175 (2024)
Graewert MA, Volkova M, Jonasson K, Määttä JAE, Gräwert T, Mamidi S, Kulesskaya N, Evenäs J, Johnsson RE, Svergun D, Bhattacharjee A, Huttunen HJ
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
75 |
nm3 |
|
|