|
|
|
|

|
| Sample: |
Adenylate cyclase toxin Block I-V monomer, 70 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl 150 mM NaCl 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Calcium-Driven Folding of RTX Domain β-Rolls Ratchets Translocation of RTX Proteins through Type I Secretion Ducts.
Mol Cell 62(1):47-62 (2016)
Bumba L, Masin J, Macek P, Wald T, Motlova L, Bibova I, Klimova N, Bednarova L, Veverka V, Kachala M, Svergun DI, Barinka C, Sebo P
|
| RgGuinier |
5.6 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Adenylate cyclase toxin Block V monomer, 16 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl 150 mM NaCl 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Calcium-Driven Folding of RTX Domain β-Rolls Ratchets Translocation of RTX Proteins through Type I Secretion Ducts.
Mol Cell 62(1):47-62 (2016)
Bumba L, Masin J, Macek P, Wald T, Motlova L, Bibova I, Klimova N, Bednarova L, Veverka V, Kachala M, Svergun DI, Barinka C, Sebo P
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Linear di-ubiquitin monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human linear tri-ubiquitin monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150mM NaCl 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human linear tetra-ubiquitin monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150mM NaCl 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
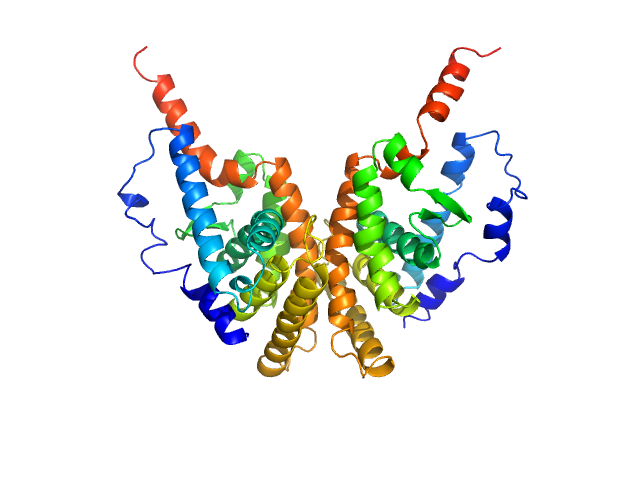
| Sample: |
Retinoic acid receptor RXR-alpha dimer, 51 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 100 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Contactin-associated protein-like 2 extracellular domains (1-1261) monomer, 140 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2010 Oct 4
|
Structural Characterization of the Extracellular Domain of CASPR2 and Insights into Its Association with the Novel Ligand Contactin1.
J Biol Chem 291(11):5788-802 (2016)
Rubio-Marrero EN, Vincelli G, Jeffries CM, Shaikh TR, Pakos IS, Ranaivoson FM, von Daake S, Demeler B, De Jaco A, Perkins G, Ellisman MH, Trewhella J, Comoletti D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
282 |
nm3 |
|
|
|
|
|
|
|
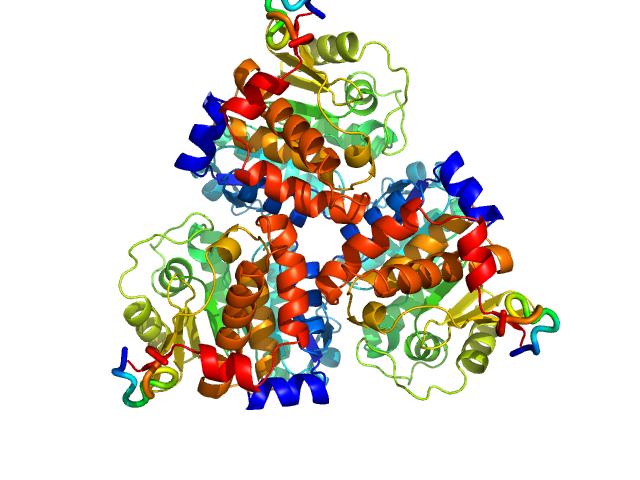
| Sample: |
Ribokinase ThiM trimer, 89 kDa Staphylococcus aureus protein
|
| Buffer: |
50 mM Potassium phosphate 10 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 19
|
Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep 6:22871 (2016)
Drebes J, Künz M, Windshügel B, Kikhney AG, Müller IB, Eberle RJ, Oberthür D, Cang H, Svergun DI, Perbandt M, Betzel C, Wrenger C
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
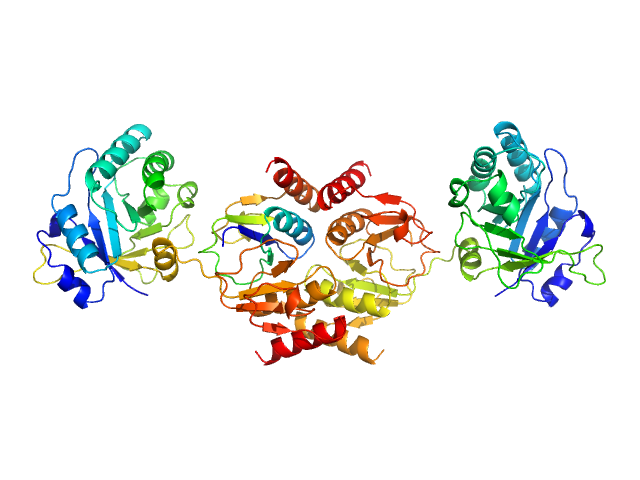
| Sample: |
Endolysin , 33 kDa Clostridium phage phiCTP1 protein
|
| Buffer: |
20 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 17
|
Crystal Structure of the CTP1L Endolysin Reveals How Its Activity Is Regulated by a Secondary Translation Product.
J Biol Chem 291(10):4882-93 (2016)
Dunne M, Leicht S, Krichel B, Mertens HD, Thompson A, Krijgsveld J, Svergun DI, Gómez-Torres N, Garde S, Uetrecht C, Narbad A, Mayer MJ, Meijers R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|
|
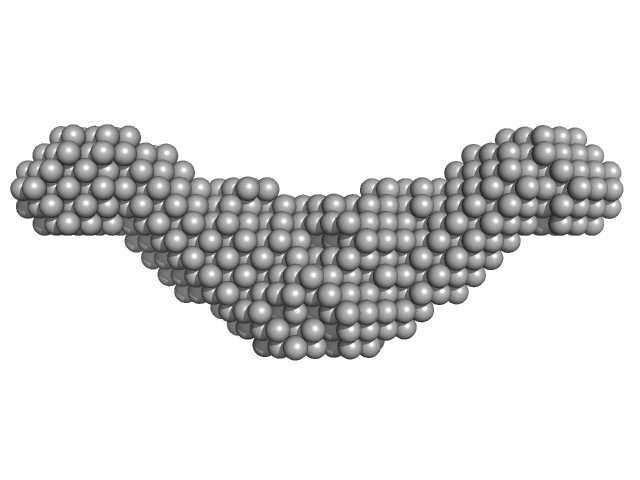
| Sample: |
Endolysin CS74L , 31 kDa Clostridium phage phi8074-B1 protein
|
| Buffer: |
20 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 17
|
Crystal Structure of the CTP1L Endolysin Reveals How Its Activity Is Regulated by a Secondary Translation Product.
J Biol Chem 291(10):4882-93 (2016)
Dunne M, Leicht S, Krichel B, Mertens HD, Thompson A, Krijgsveld J, Svergun DI, Gómez-Torres N, Garde S, Uetrecht C, Narbad A, Mayer MJ, Meijers R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
79 |
nm3 |
|
|