|
|
|
|
|
| Sample: |
Ethylene Receptor 1 dimer, 129 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM TRIS 150 mM NaCl 1mM DTT 250mM NDSB, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 29
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
5.5 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ethylene receptor 1 dimerization histidine phosphotransfer + catalytic ATP-binding + receiver domains dimer, 88 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM DTT 5 mM ADP, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 11
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
144 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ethylene receptor 1 dimerization histidine phosphotransfer + catalytic ATP-binding domains dimer, 55 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM DTT 5 mM ADP, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 11
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
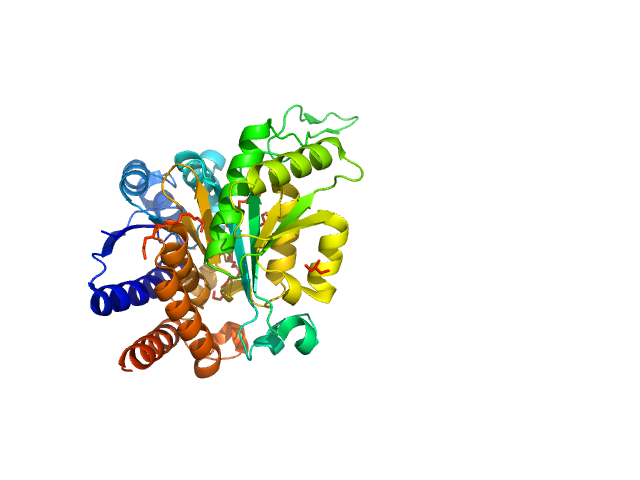
|
| Sample: |
3-isopropylmalate dehydrogenase dimer, 73 kDa Thermus thermophilus protein
|
| Buffer: |
25 mM MOPS/NaOH, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 3
|
Glutamate 270 plays an essential role in K(+)-activation and domain closure of Thermus thermophilus isopropylmalate dehydrogenase.
FEBS Lett 589(2):240-5 (2015)
Gráczer É, Palló A, Oláh J, Szimler T, Konarev PV, Svergun DI, Merli A, Závodszky P, Weiss MS, Vas M
|
|
|
|
|
|
|
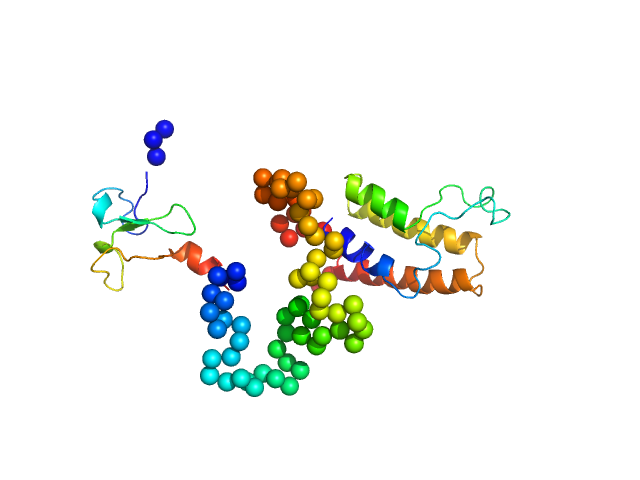
|
| Sample: |
Bromodomain adjacent to zinc finger domain protein 2A monomer, 27 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 500 mM NaCl 2 mM DTT 10 microM ZnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 16
|
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 23(1):80-92 (2015)
Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
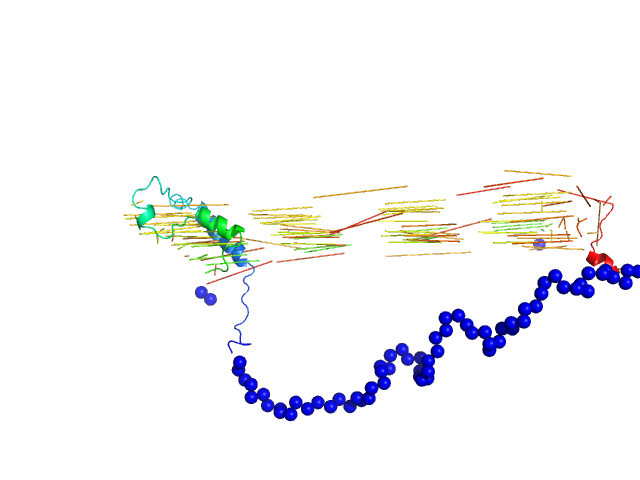
|
| Sample: |
Bromodomain adjacent to zinc finger domain protein 2B, C-terminal monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 500 mM NaCl 2 mM DTT 10 microM ZnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 24
|
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 23(1):80-92 (2015)
Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bromodomain adjacent to zinc finger domain protein 2B, C-terminal monomer, 28 kDa Homo sapiens protein
H3Kac9Kac14 monomer, 5 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 500 mM NaCl 2 mM DTT 10 microM ZnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 24
|
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 23(1):80-92 (2015)
Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
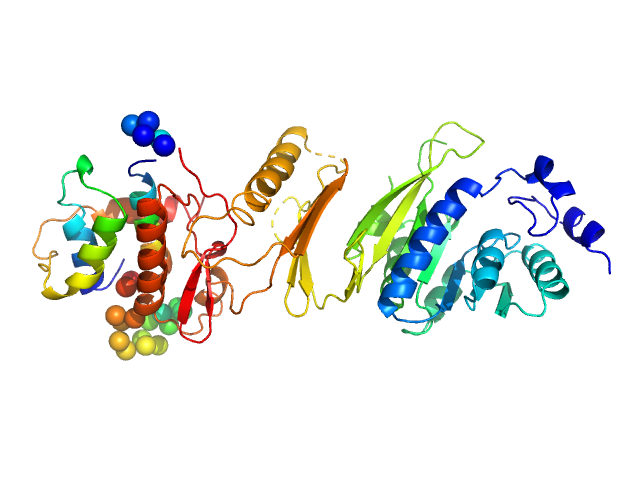
|
| Sample: |
Bifunctional kinase- methyltransferase WbdD monomer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM BisTris 50 mM NaCl 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Sep 23
|
A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide.
Nat Struct Mol Biol 22(1):50-56 (2015)
Hagelueken G, Clarke BR, Huang H, Tuukkanen A, Danciu I, Svergun DI, Hussain R, Liu H, Whitfield C, Naismith JH
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
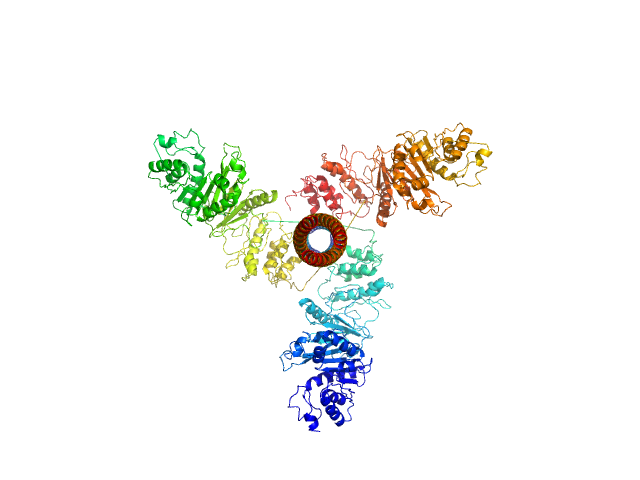
| Sample: |
Bifunctional kinase- methyltransferase WbdD trimer, 190 kDa protein
|
| Buffer: |
20 mM BisTris 50 mM NaCl 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 2
|
A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide.
Nat Struct Mol Biol 22(1):50-56 (2015)
Hagelueken G, Clarke BR, Huang H, Tuukkanen A, Danciu I, Svergun DI, Hussain R, Liu H, Whitfield C, Naismith JH
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human Filamin A Ig-like domains 20-21/migfilin peptide complex monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 1
|
Flexible Structure of Peptide-Bound Filamin A Mechanosensor Domain Pair 20-21.
PLoS One 10(8):e0136969 (2015)
Seppälä J, Tossavainen H, Rodic N, Permi P, Pentikäinen U, Ylänne J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
31 |
nm3 |
|
|